How to develop a meaningful radiomic signature for clinical use in oncologic patients
- PMID: 32357923
- PMCID: PMC7195800
- DOI: 10.1186/s40644-020-00311-4
How to develop a meaningful radiomic signature for clinical use in oncologic patients
Abstract
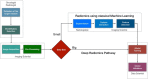
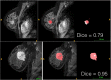
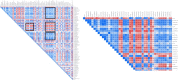
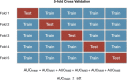
During the last decade, there is an increasing usage of quantitative methods in Radiology in an effort to reduce the diagnostic variability associated with a subjective manner of radiological interpretation. Combined approaches where visual assessment made by the radiologist is augmented by quantitative imaging biomarkers are gaining attention. Advances in machine learning resulted in the rise of radiomics that is a new methodology referring to the extraction of quantitative information from medical images. Radiomics are based on the development of computational models, referred to as "Radiomic Signatures", trying to address either unmet clinical needs, mostly in the field of oncologic imaging, or to compare radiomics performance with that of radiologists. However, to explore this new technology, initial publications did not consider best practices in the field of machine learning resulting in publications with questionable clinical value. In this paper, our effort was concentrated on how to avoid methodological mistakes and consider critical issues in the workflow of the development of clinically meaningful radiomic signatures.
Keywords: Machine learning; Quantitative imaging; Radiomics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

