BRCA1 Mutational Complementation Induces Synthetic Viability
- PMID: 32359443
- PMCID: PMC7418109
- DOI: 10.1016/j.molcel.2020.04.006
BRCA1 Mutational Complementation Induces Synthetic Viability
Abstract
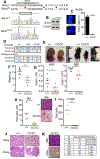
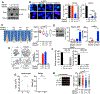
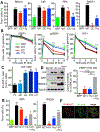
BRCA1 promotes the DNA end resection and RAD51 loading steps of homologous recombination (HR). Whether these functions can be uncoupled, and whether mutant proteins retaining partial activity can complement one another, is unclear and could affect the severity of BRCA1-associated Fanconi anemia (FA). Here we generated a Brca1CC mouse with a coiled-coil (CC) domain deletion. Brca1CC/CC mice are born at low frequencies, and post-natal mice have FA-like abnormalities, including bone marrow failure. Intercrossing with Brca1Δ11, which is homozygous lethal, generated Brca1CC/Δ11 mice at Mendelian frequencies that were indistinguishable from Brca1+/+ mice. Brca1CC and Brca1Δ11 proteins were individually responsible for counteracting 53BP1-RIF1-Shieldin activity and promoting RAD51 loading, respectively. Thus, Brca1CC and Brca1Δ11 alleles represent separation-of-function mutations that combine to provide a level of HR sufficient for normal development and hematopoiesis. Because BRCA1 activities can be genetically separated, compound heterozygosity for functional complementary mutations may protect individuals from FA.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no conflicts of interest.
Figures

References
-
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, and Helleday T (2005). Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

