Activity-Based Diagnostics: An Emerging Paradigm for Disease Detection and Monitoring
- PMID: 32359477
- PMCID: PMC8290463
- DOI: 10.1016/j.molmed.2020.01.013
Activity-Based Diagnostics: An Emerging Paradigm for Disease Detection and Monitoring
Abstract
Diagnostics to accurately detect disease and monitor therapeutic response are essential for effective clinical management. Bioengineering, chemical biology, molecular biology, and computer science tools are converging to guide the design of diagnostics that leverage enzymatic activity to measure or produce biomarkers of disease. We review recent advances in the development of these 'activity-based diagnostics' (ABDx) and their application in infectious and noncommunicable diseases. We highlight efforts towards both molecular probes that respond to disease-specific catalytic activity to produce a diagnostic readout, as well as diagnostics that use enzymes as an engineered component of their sense-and-respond cascade. These technologies exemplify how integrating techniques from multiple disciplines with preclinical validation has enabled ABDx that may realize the goals of precision medicine.
Keywords: CRISPR/Cas; activity-based probes; diagnostics; enzymes; protease activity; synthetic biology; synthetic biomarkers.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
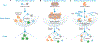
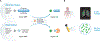



References
-
- Etzioni R et al. (2003) The case for early detection. Nat. Rev. Cancer 3, 243–252 - PubMed
-
- Siegel RL et al. (2019) Cancer statistics, 2019. CA. Cancer J. Clin 69, 7–34 - PubMed
-
- American Thoracic Society (2001) Guidelines for the management of adults with community-acquired pneumonia. Am. J. Respir. Crit. Care Med 163, 1730–1754 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

