Oligomerization of a symmetric β-trefoil protein in response to folding nucleus perturbation
- PMID: 32362013
- PMCID: PMC7314399
- DOI: 10.1002/pro.3877
Oligomerization of a symmetric β-trefoil protein in response to folding nucleus perturbation
Abstract
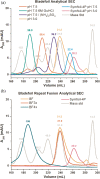
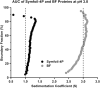
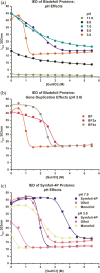
Gene duplication and fusion events in protein evolution are postulated to be responsible for the common protein folds exhibiting internal rotational symmetry. Such evolutionary processes can also potentially yield regions of repetitive primary structure. Repetitive primary structure offers the potential for alternative definitions of critical regions, such as the folding nucleus (FN). In principle, more than one instance of the FN potentially enables an alternative folding pathway in the face of a subsequent deleterious mutation. We describe the targeted mutation of the carboxyl-terminal region of the (internally located) FN of the de novo designed purely-symmetric β-trefoil protein Symfoil-4P. This mutation involves wholesale replacement of a repeating trefoil-fold motif with a "blade" motif from a β-propeller protein, and postulated to trap that region of the Symfoil-4P FN in a nonproductive folding intermediate. The resulting protein (termed "Bladefoil") is shown to be cooperatively folding, but as a trimeric oligomer. The results illustrate how symmetric protein architectures have potentially diverse folding alternatives available to them, including oligomerization, when preferred pathways are perturbed.
Keywords: domain-swapping; folding pathway; protein evolution; protein symmetry.
© 2020 The Protein Society.
Conflict of interest statement
M. B. is a cofounder and has equity ownership in Trefoil Therapeutics, Inc.
Figures
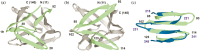




References
-
- McLachlan AD. Three‐fold structural pattern in the soybean trypsin inhibitor (Kunitz). J Mol Biol. 1979;133:557–563. - PubMed
-
- Murzin AG, Lesk AM, Chothia C. β‐Trefoil fold. Patterns of structure and sequence in the kunitz inhibitors interleukins‐1β and 1α and fibroblast growth factors. J Mol Biol. 1992;223:531–543. - PubMed
-
- Lee J, Blaber SI, Dubey VK, Blaber M. A polypeptide “building block” for the ß‐trefoil fold identified by “top‐down symmetric deconstruction”. J Mol Biol. 2011;407:744–763. - PubMed
-
- Broom A, Doxey AC, Lobsanov YD, et al. Modular evolution and the origins of symmetry: Reconstruction of a three‐fold symmetric globular protein. Structure. 2012;20:1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

