Exploring the Potential of the Microbiome as a Marker of the Geographic Origin of Fresh Seafood
- PMID: 32362885
- PMCID: PMC7181054
- DOI: 10.3389/fmicb.2020.00696
Exploring the Potential of the Microbiome as a Marker of the Geographic Origin of Fresh Seafood
Abstract
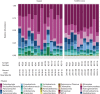
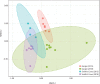
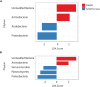
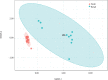
Geographic food fraud - misrepresenting the geographic origin of a food item, is very difficult to detect, and therefore this type of fraud tends to go undetected. This potentially negatively impacts the health of Canadians and economic success of our seafood industry. Surveillance studies have shown that up to a significant portion of commercially sold seafood items in Canada are mislabeled or otherwise misrepresented in some way. The current study aimed to determine if the microbiome of fresh shellfish could be used as an accurate marker of harvest location. Total DNA was extracted from the homogenate of 25 batches of fresh soft-shell clams (Mya arenaria) harvested in 2015 and 2018 from two locations on the East Coast of Canada and the microbiome of each homogenate was characterized using 16S rRNA targeted amplicon sequencing. Clams harvested from Nova Scotia in both years had a higher abundance of Proteobacteria and Acidobacteria (p < 0.05), but a lower abundance of Actinobacteria (p < 0.05) than those from Quebec. Alpha-diversity also differed significantly between sites. Samples harvested from Nova Scotia had greater diversity (p < 0.0001) than those from Quebec. Beta-diversity analysis showed that the microbial community composition was significantly different between the samples from Nova Scotia and Quebec and indicated that 16S rRNA targeted amplicon sequencing might be an effective tool for elucidating the geographic origin of unprocessed shellfish. To evaluate if the microbiome of shellfish experiences a loss of microbial diversity during processing and storage - which would limit the ability of this technique to link retail samples to geographic origin, 10 batches of retail clams purchased from grocery stores were also examined. Microbial diversity and species richness was significantly lower in retail clams, and heavily dominated by Proteobacteria, a typical spoilage organism for fresh seafood, this may make determining the geographic origin of seafood items more difficult in retail clams than in freshly harvested clams.
Keywords: adulteration and substitution; food fraud prevention; microbiome; mothur; seafood.
Copyright © 2020 Liu, Teixeira, Ner, Ma, Petronella, Banerjee and Ronholm.
Figures
References
-
- BC Centre for Disease Control (2019). Shellfish Safety Notes. Available online at: http://www.bccdc.ca/resource-gallery/Documents/Educational%20Materials/E... (accessed October 24, 2019).
-
- Black C., Chevallier O. P., Elliott C. T. (2016). The current and potential applications of Ambient Mass Spectrometry in detecting food fraud. TrAC Trends Anal. Chem. 82 268–278. 10.1016/j.trac.2016.06.005 - DOI
-
- Chatterjee N. S., Chevallier O. P., Wielogorska E., Black C., Elliott C. T. (2019). Simultaneous authentication of species identity and geographical origin of shrimps: Untargeted metabolomics to recurrent biomarker ions. J. Chromatogr. A 1599 75–84. - PubMed
-
- Chen K. L. A., Liu X., Zhao Y. C., Hieronymi K., Rossi G., Auvil L. S., et al. (2018). Long-term administration of conjugated estrogen and bazedoxifene decreased murine fecal β-glucuronidase activity without impacting overall microbiome community. Sci. Rep. 8:8166. 10.1038/s41598-018-26506-1 - DOI - PMC - PubMed
-
- Christoph N., Hermann A., Wachter H. (2015). 25 Years authentication of wine with stable isotope analysis in the European Union – Review and outlook. BIO Web. Confer. 5:02020 10.1051/bioconf/20150502020 - DOI
LinkOut - more resources
Full Text Sources

