Optimized Whole Genome Association Scanning for Discovery of HLA Class I-Restricted Minor Histocompatibility Antigens
- PMID: 32362897
- PMCID: PMC7180171
- DOI: 10.3389/fimmu.2020.00659
Optimized Whole Genome Association Scanning for Discovery of HLA Class I-Restricted Minor Histocompatibility Antigens
Abstract
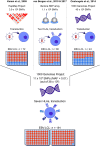
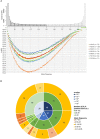
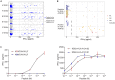
Patients undergoing allogeneic stem cell transplantation as treatment for hematological diseases face the risk of Graft-versus-Host Disease as well as relapse. Graft-versus-Host Disease and the favorable Graft-versus-Leukemia effect are mediated by donor T cells recognizing polymorphic peptides, which are presented on the cell surface by HLA molecules and result from single nucleotide polymorphism alleles that are disparate between patient and donor. Identification of polymorphic HLA-binding peptides, designated minor histocompatibility antigens, has been a laborious procedure, and the number and scope for broad clinical use of these antigens therefore remain limited. Here, we present an optimized whole genome association approach for discovery of HLA class I minor histocompatibility antigens. T cell clones isolated from patients who responded to donor lymphocyte infusions after HLA-matched allogeneic stem cell transplantation were tested against a panel of 191 EBV-transformed B cells, which have been sequenced by the 1000 Genomes Project and selected for expression of seven common HLA class I alleles (HLA-A∗01:01, A∗02:01, A∗03:01, B∗07:02, B∗08:01, C∗07:01, and C∗07:02). By including all polymorphisms with minor allele frequencies above 0.01, we demonstrated that the new approach allows direct discovery of minor histocompatibility antigens as exemplified by seven new antigens in eight different HLA class I alleles including one antigen in HLA-A∗24:02 and HLA-A∗23:01, for which the method has not been originally designed. Our new whole genome association strategy is expected to rapidly augment the repertoire of HLA class I-restricted minor histocompatibility antigens that will become available for donor selection and clinical use to predict, follow or manipulate Graft-versus-Leukemia effect and Graft-versus-Host Disease after allogeneic stem cell transplantation.
Keywords: Graft-versus-Leukemia effect; HLA class I; allogeneic stem cell transplantation; graft versus host disease; hematological diseases; minor histocompatibility antigens; whole genome association scanning.
Copyright © 2020 Fuchs, Honders, van der Meijden, Adriaans, van der Lee, Pont, Monajemi, Kielbasa, ’t Hoen, van Bergen, Falkenburg and Griffioen.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

