Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2
- PMID: 32364527
- PMCID: PMC7244084
- DOI: 10.18632/aging.103101
Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2
Abstract
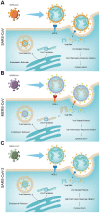
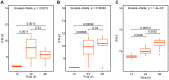
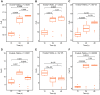
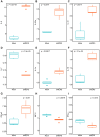
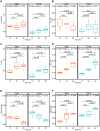
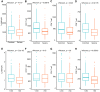
Currently, we are on a global pandemic of Coronavirus disease-2019 (COVID-19) which causes fever, dry cough, fatigue and acute respiratory distress syndrome (ARDS) that may ultimately lead to the death of the infected. Current researches on COVID-19 continue to highlight the necessity for further understanding the virus-host synergies. In this study, we have highlighted the key cytokines induced by coronavirus infections. We have demonstrated that genes coding interleukins (Il-1α, Il-1β, Il-6, Il-10), chemokine (Ccl2, Ccl3, Ccl5, Ccl10), and interferon (Ifn-α2, Ifn-β1, Ifn2) upsurge significantly which in line with the elevated infiltration of T cells, NK cells and monocytes in SARS-Cov treated group at 24 hours. Also, interleukins (IL-6, IL-23α, IL-10, IL-7, IL-1α, IL-1β) and interferon (IFN-α2, IFN2, IFN-γ) have increased dramatically in MERS-Cov at 24 hours. A similar cytokine profile showed the cytokine storm served a critical role in the infection process. Subsequent investigation of 463 patients with COVID-19 disease revealed the decreased amount of total lymphocytes, CD3+, CD4+, and CD8+ T lymphocytes in the severe type patients which indicated COVID-19 can impose hard blows on human lymphocyte resulting in lethal pneumonia. Thus, taking control of changes in immune factors could be critical in the treatment of COVID-19.
Keywords: COVID-19; SARS-Cov-2; cytokine storm.
Conflict of interest statement
Figures


Similar articles
-
Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection.Front Immunol. 2020 May 12;11:1022. doi: 10.3389/fimmu.2020.01022. eCollection 2020. Front Immunol. 2020. PMID: 32574260 Free PMC article. Review.
-
Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past.Front Immunol. 2020 Aug 7;11:1949. doi: 10.3389/fimmu.2020.01949. eCollection 2020. Front Immunol. 2020. PMID: 32849654 Free PMC article. Review.
-
Innate Immune Responses to Highly Pathogenic Coronaviruses and Other Significant Respiratory Viral Infections.Front Immunol. 2020 Aug 18;11:1979. doi: 10.3389/fimmu.2020.01979. eCollection 2020. Front Immunol. 2020. PMID: 32973803 Free PMC article. Review.
-
Overview of lethal human coronaviruses.Signal Transduct Target Ther. 2020 Jun 10;5(1):89. doi: 10.1038/s41392-020-0190-2. Signal Transduct Target Ther. 2020. PMID: 32533062 Free PMC article. Review.
-
Cyclosporine therapy during the COVID-19 pandemic.J Am Acad Dermatol. 2020 Aug;83(2):e151-e152. doi: 10.1016/j.jaad.2020.04.153. Epub 2020 May 4. J Am Acad Dermatol. 2020. PMID: 32376422 Free PMC article. No abstract available.
Cited by
-
SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes.Cell Death Discov. 2021 Mar 1;7(1):43. doi: 10.1038/s41420-021-00428-w. Cell Death Discov. 2021. PMID: 33649297 Free PMC article.
-
Identification of COVID-19 and Dengue Host Factor Interaction Networks Based on Integrative Bioinformatics Analyses.Front Immunol. 2021 Jul 28;12:707287. doi: 10.3389/fimmu.2021.707287. eCollection 2021. Front Immunol. 2021. PMID: 34394108 Free PMC article.
-
Factors associated with disease severity and mortality among patients with COVID-19: A systematic review and meta-analysis.PLoS One. 2020 Nov 18;15(11):e0241541. doi: 10.1371/journal.pone.0241541. eCollection 2020. PLoS One. 2020. PMID: 33206661 Free PMC article.
-
A Missing Link: Engagements of Dendritic Cells in the Pathogenesis of SARS-CoV-2 Infections.Int J Mol Sci. 2021 Jan 23;22(3):1118. doi: 10.3390/ijms22031118. Int J Mol Sci. 2021. PMID: 33498725 Free PMC article. Review.
-
Immune modulation: the key to combat SARS-CoV-2 induced myocardial injury.Front Immunol. 2025 May 14;16:1561946. doi: 10.3389/fimmu.2025.1561946. eCollection 2025. Front Immunol. 2025. PMID: 40438117 Free PMC article. Review.
References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, et al., and SARS Working Group. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003; 348:1953–66. 10.1056/NEJMoa030781 - DOI - PubMed
-
- de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S, Poon LL, Snijder EJ, et al.. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013; 87:7790–92. 10.1128/JVI.01244-13 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

