Tuning antiviral CD8 T-cell response via proline-altered peptide ligand vaccination
- PMID: 32365082
- PMCID: PMC7224568
- DOI: 10.1371/journal.ppat.1008244
Tuning antiviral CD8 T-cell response via proline-altered peptide ligand vaccination
Abstract
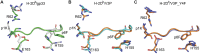
Viral escape from CD8+ cytotoxic T lymphocyte responses correlates with disease progression and represents a significant challenge for vaccination. Here, we demonstrate that CD8+ T cell recognition of the naturally occurring MHC-I-restricted LCMV-associated immune escape variant Y4F is restored following vaccination with a proline-altered peptide ligand (APL). The APL increases MHC/peptide (pMHC) complex stability, rigidifies the peptide and facilitates T cell receptor (TCR) recognition through reduced entropy costs. Structural analyses of pMHC complexes before and after TCR binding, combined with biophysical analyses, revealed that although the TCR binds similarly to all complexes, the p3P modification alters the conformations of a very limited amount of specific MHC and peptide residues, facilitating efficient TCR recognition. This approach can be easily introduced in peptides restricted to other MHC alleles, and can be combined with currently available and future vaccination protocols in order to prevent viral immune escape.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Achour A, Michaelsson J, Harris RA, Odeberg J, Grufman P, Sandberg JK, et al. A structural basis for LCMV immune evasion: subversion of H-2D(b) and H-2K(b) presentation of gp33 revealed by comparative crystal structure.Analyses. Immunity. 2002;17(6):757–68. 10.1016/s1074-7613(02)00478-8 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

