Leukamenin E Induces K8/18 Phosphorylation and Blocks the Assembly of Keratin Filament Networks Through ERK Activation
- PMID: 32365802
- PMCID: PMC7246489
- DOI: 10.3390/ijms21093164
Leukamenin E Induces K8/18 Phosphorylation and Blocks the Assembly of Keratin Filament Networks Through ERK Activation
Abstract
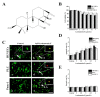
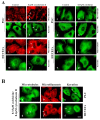
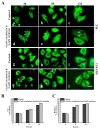
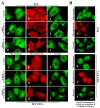
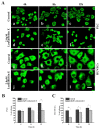
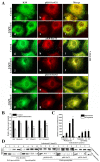
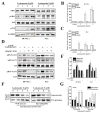
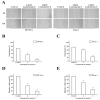
Leukamenin E is a natural ent-kaurane diterpenoid isolated from Isodon racemosa (Hemsl) Hara that has been found to be a novel and potential keratin filament inhibitor, but its underlying mechanisms remain largely unknown. Here, we show that leukamenin E induces keratin filaments (KFs) depolymerization, largely independently of microfilament (MFs) and microtubules (MTs) in well-spread cells and inhibition of KFs assembly in spreading cells. These effects are accompanied by keratin phosphorylation at K8-Ser73/Ser431 and K18-Ser52 via the by extracellular signal-regulated kinases (ERK) pathway in primary liver carcinoma cells (PLC) and human umbilical vein endothelial cells (HUVECs). Moreover, leukamenin E increases soluble pK8-Ser73/Ser431, pK18-Ser52, and pan-keratin in the cytoplasmic supernatant by immunofluorescence imaging and Western blotting assay. Accordingly, leukamenin E inhibits the spreading and migration of cells. We propose that leukamenin E-induced keratin phosphorylation may interfere with the initiation of KFs assembly and block the formation of a new KFs network, leading to the inhibition of cell spreading. Leukamenin E is a potential target drug for inhibition of KFs assembly.
Keywords: inhibitory effect of keratin filaments assembly 3; leukamenin E 1; phosphorylation of keratin 2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hesse M., Magin T.M., Weber K. Genes for intermediate filament proteins and the draft sequence of the human genome: Novel keratin genes and a surprisingly high number of pseudogenes related to keratin genes 8 and 18. Cell Sci. 2001;114:2569–2575. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

