A collection of genetic mouse lines and related tools for inducible and reversible intersectional mis-expression
- PMID: 32366677
- PMCID: PMC7272339
- DOI: 10.1242/dev.186650
A collection of genetic mouse lines and related tools for inducible and reversible intersectional mis-expression
Abstract
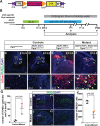
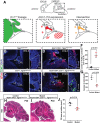
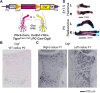
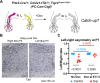
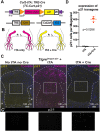
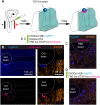
Thanks to many advances in genetic manipulation, mouse models have become very powerful in their ability to interrogate biological processes. In order to precisely target expression of a gene of interest to particular cell types, intersectional genetic approaches using two promoter/enhancers unique to a cell type are ideal. Within these methodologies, variants that add temporal control of gene expression are the most powerful. We describe the development, validation and application of an intersectional approach that involves three transgenes, requiring the intersection of two promoter/enhancers to target gene expression to precise cell types. Furthermore, the approach uses available lines expressing tTA/rTA to control the timing of gene expression based on whether doxycycline is absent or present, respectively. We also show that the approach can be extended to other animal models, using chicken embryos. We generated three mouse lines targeted at the Tigre (Igs7) locus with TRE-loxP-tdTomato-loxP upstream of three genes (p21, DTA and Ctgf), and combined them with Cre and tTA/rtTA lines that target expression to the cerebellum and limbs. Our tools will facilitate unraveling biological questions in multiple fields and organisms.
Keywords: Ccn2/Ctgf; Cre; DRAGON; DTA; Diphtheria toxin; Inducible and reversible gene mis-expression; Intersectional genetics; Tigre locus; Tissue-specific; p21; rtTA; tTA; tox176.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




References
-
- Belteki G., Haigh J., Kabacs N., Haigh K., Sison K., Costantini F., Whitsett J., Quaggin S. E. and Nagy A. (2005). Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 33, e51 10.1093/nar/gni051 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

