Intracellular Staphylococcus aureus persisters upon antibiotic exposure
- PMID: 32366839
- PMCID: PMC7198484
- DOI: 10.1038/s41467-020-15966-7
Intracellular Staphylococcus aureus persisters upon antibiotic exposure
Abstract
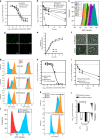
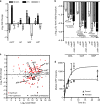
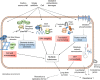
Bacterial persister cells are phenotypic variants that exhibit a transient non-growing state and antibiotic tolerance. Here, we provide in vitro evidence of Staphylococcus aureus persisters within infected host cells. We show that the bacteria surviving antibiotic treatment within host cells are persisters, displaying biphasic killing and reaching a uniformly non-responsive, non-dividing state when followed at the single-cell level. This phenotype is stable but reversible upon antibiotic removal. Intracellular S. aureus persisters remain metabolically active but display an altered transcriptomic profile consistent with activation of stress responses, including the stringent response as well as cell wall stress, SOS and heat shock responses. These changes are associated with multidrug tolerance after exposure to a single antibiotic. We hypothesize that intracellular S. aureus persisters may constitute a reservoir for relapsing infection and could contribute to therapeutic failures.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. doi: 10.1016/S0140-6736(00)74210-3. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

