Transcriptional and epigenetic basis of Treg cell development and function: its genetic anomalies or variations in autoimmune diseases
- PMID: 32367041
- PMCID: PMC7264322
- DOI: 10.1038/s41422-020-0324-7
Transcriptional and epigenetic basis of Treg cell development and function: its genetic anomalies or variations in autoimmune diseases
Abstract
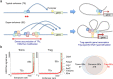
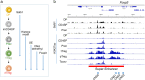
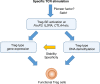
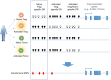
Naturally arising regulatory CD4+ T (Treg) cells, which specifically express the transcription factor FoxP3 in the nucleus and CD25 and CTLA-4 on the cell surface, are a T-cell subpopulation specialized for immune suppression, playing a key role in maintaining immunological self-tolerance and homeostasis. FoxP3 is required for Treg function, especially for its suppressive activity. However, FoxP3 expression per se is not necessary for Treg cell lineage commitment in the thymus and insufficient for full Treg-type gene expression in mature Treg cells. It is Treg-specific epigenetic changes such as CpG demethylation and histone modification that can confer a stable and heritable pattern of Treg type gene expression on developing Treg cells in a FoxP3-independent manner. Anomalies in the formation of Treg-specific epigenome, in particular, Treg-specific super-enhancers, which largely include Treg-specific DNA demethylated regions, are indeed able to cause autoimmune diseases in rodents. Furthermore, in humans, single nucleotide polymorphisms in Treg-specific DNA demethylated regions associated with Treg signature genes, such as IL2RA (CD25) and CTLA4, can affect the development and function of naïve Treg cells rather than effector T cells. Such genetic variations are therefore causative of polygenic common autoimmune diseases including type 1 diabetes and rheumatoid arthritis via affecting endogenous natural Treg cells. These findings on the transcription factor network with FoxP3 at a key position as well as Treg-specific epigenetic landscape facilitate our understanding of Treg cell development and function, and can be exploited to prepare functionally stable FoxP3-expressing Treg cells from antigen-specific conventional T cells to treat autoimmune diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. - PubMed
-
- Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 2004;22:531–562. - PubMed
-
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–1164. - PubMed
-
- Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

