FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome
- PMID: 32367049
- PMCID: PMC7286794
- DOI: 10.1038/s41556-020-0511-2
FBP1 loss disrupts liver metabolism and promotes tumorigenesis through a hepatic stellate cell senescence secretome
Abstract
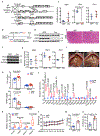
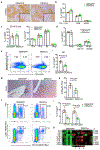
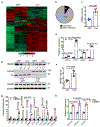
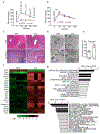
The crosstalk between deregulated hepatocyte metabolism and cells within the tumour microenvironment, as well as the consequent effects on liver tumorigenesis, are not completely understood. We show here that hepatocyte-specific loss of the gluconeogenic enzyme fructose 1,6-bisphosphatase 1 (FBP1) disrupts liver metabolic homeostasis and promotes tumour progression. FBP1 is universally silenced in both human and murine liver tumours. Hepatocyte-specific Fbp1 deletion results in steatosis, concomitant with activation and senescence of hepatic stellate cells (HSCs), exhibiting a senescence-associated secretory phenotype. Depleting senescent HSCs by 'senolytic' treatment with dasatinib/quercetin or ABT-263 inhibits tumour progression. We further demonstrate that FBP1-deficient hepatocytes promote HSC activation by releasing HMGB1; blocking its release with the small molecule inflachromene limits FBP1-dependent HSC activation, the subsequent development of the senescence-associated secretory phenotype and tumour progression. Collectively, these findings provide genetic evidence for FBP1 as a metabolic tumour suppressor in liver cancer and establish a critical crosstalk between hepatocyte metabolism and HSC senescence that promotes tumour growth.
Conflict of interest statement
Competing interests
The authors declare no competing financial interests.
Figures











Comment in
-
Hepatic Stellate Cell Senescence in Liver Tumorigenesis.Hepatology. 2021 Feb;73(2):853-855. doi: 10.1002/hep.31556. Hepatology. 2021. PMID: 32931043 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

