CDK11 is required for transcription of replication-dependent histone genes
- PMID: 32367068
- PMCID: PMC7116321
- DOI: 10.1038/s41594-020-0406-8
CDK11 is required for transcription of replication-dependent histone genes
Abstract
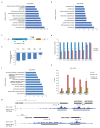
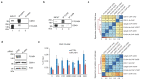
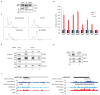
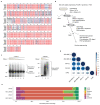
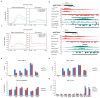
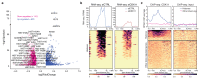
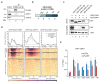
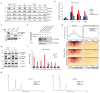
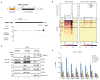
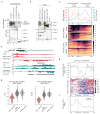
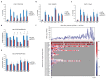
Replication-dependent histones (RDH) are required for packaging of newly synthetized DNA into nucleosomes during the S phase when their expression is highly upregulated. However, the mechanisms of this upregulation in metazoan cells remain poorly understood. Using iCLIP and ChIP-seq, we found that human cyclin-dependent kinase 11 (CDK11) associates with RNA and chromatin of RDH genes primarily in the S phase. Moreover, its amino-terminal region binds FLASH, an RDH-specific 3'-end processing factor, which keeps the kinase on the chromatin. CDK11 phosphorylates serine 2 (Ser2) of the carboxy-terminal domain of RNA polymerase II (RNAPII), which is initiated when RNAPII reaches the middle of RDH genes and is required for further RNAPII elongation and 3'-end processing. CDK11 depletion leads to decreased number of cells in S phase, likely owing to the function of CDK11 in RDH gene expression. Thus, the reliance of RDH expression on CDK11 could explain why CDK11 is essential for the growth of many cancers.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

