Long Non-Coding RNA-NEAT1 Promotes Cell Migration and Invasion via Regulating miR-124/NF-κB Pathway in Cervical Cancer
- PMID: 32368085
- PMCID: PMC7173957
- DOI: 10.2147/OTT.S220306
Long Non-Coding RNA-NEAT1 Promotes Cell Migration and Invasion via Regulating miR-124/NF-κB Pathway in Cervical Cancer
Abstract
Background: This study aimed to investigate the regulatory role of lncRNA-NEAT1 on cervical cancer (CC) and the underlying molecular mechanisms.
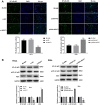
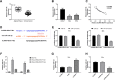
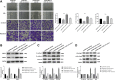
Methods: The expression of lncRNA-NEAT1 and miR-124 was detected in CC tissues and cells (HeLa and SiHa cells) by qRT-RCR. The relation between lncRNA-NEAT1 expression and clinical parameters of CC patients was explored. The cell migration and invasion were detected by wound healing assay and transwell assay. The cell proliferation was detected by CCK-8 and anchorage-independent colony assay. The targeting relation between miR-124 and lncRNA-NEAT1 was predicted by TargetScan and identified by dual luciferase reporter gene and RNA pull-down assay. The expression of metastasis- (MMP-2 and MMP), EMT- (E-cadherin, N-cadherin and Vimentin), and NF-κB pathway-related factors (NF-κB p65, p-NF-κB p65 and IκBα) was detected by Western blot.
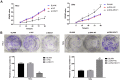
Results: The expression of lncRNA-NEAT1 was upregulated in CC tissues and cells and positively correlated with TNM stage and lymph node metastasis. Overexpression of lncRNA-NEAT1 promoted the proliferation, migration and invasion, influenced the expression of EMT markers, and activated NF-κB pathway in HeLa and SiHa cells. Silencing of lncRNA-NEAT1 exhibited opposite effects on HeLa and SiHa cells. LncRNA-NEAT1 could negatively regulate its target miR-124. MiR-124 reversed the effects of lncRNA-NEAT1 on the migration, invasion, EMT and NF-κB pathway of HeLa cells.
Conclusion: LncRNA-NEAT1 promoted the migration and invasion of CC cells via regulating miR-124/NF-κB pathway.
Keywords: cervical cancer; invasion; lncRNA-NEAT1; miR-124/NF-κB pathway; migration.
© 2020 Shen et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures



Similar articles
-
NF-κB interaction long non-coding RNA inhibits migration, invasion and epithelial-mesenchymal transition of cervical cancer cells through inhibiting NF-κB signaling pathways.Exp Ther Med. 2020 Aug;20(2):1039-1047. doi: 10.3892/etm.2020.8752. Epub 2020 May 14. Exp Ther Med. 2020. PMID: 32765657 Free PMC article.
-
Long Non-Coding RNA NEAT1 Promotes the Proliferation, Migration, and Metastasis of Human Breast-Cancer Cells by Inhibiting miR-146b-5p Expression.Cancer Manag Res. 2020 Jul 21;12:6091-6101. doi: 10.2147/CMAR.S252295. eCollection 2020. Cancer Manag Res. 2020. PMID: 32801860 Free PMC article.
-
Knockdown of lncRNA NEAT1 expression inhibits cell migration, invasion and EMT by regulating the miR-24-3p/LRG1 axis in retinoblastoma cells.Exp Ther Med. 2021 Apr;21(4):367. doi: 10.3892/etm.2021.9798. Epub 2021 Feb 18. Exp Ther Med. 2021. PMID: 33732340 Free PMC article.
-
The role of lncRNA NEAT1 in human cancer chemoresistance.Cancer Cell Int. 2024 Jul 5;24(1):236. doi: 10.1186/s12935-024-03426-x. Cancer Cell Int. 2024. PMID: 38970092 Free PMC article. Review.
-
Mechanism of action of lncRNA-NEAT1 in immune diseases.Front Genet. 2025 Mar 5;16:1501115. doi: 10.3389/fgene.2025.1501115. eCollection 2025. Front Genet. 2025. PMID: 40110044 Free PMC article. Review.
Cited by
-
The role of NF-κB in carcinogenesis of cervical cancer: opportunities and challenges.Mol Biol Rep. 2024 Apr 20;51(1):538. doi: 10.1007/s11033-024-09447-z. Mol Biol Rep. 2024. PMID: 38642209 Review.
-
In Silico Identification of Potential Quadruplex Forming Sequences in LncRNAs of Cervical Cancer.Int J Mol Sci. 2023 Aug 10;24(16):12658. doi: 10.3390/ijms241612658. Int J Mol Sci. 2023. PMID: 37628839 Free PMC article.
-
The Role of Long Non-Coding RNAs (lncRNAs) in Female Oriented Cancers.Cancers (Basel). 2021 Dec 3;13(23):6102. doi: 10.3390/cancers13236102. Cancers (Basel). 2021. PMID: 34885213 Free PMC article. Review.
-
The associations and roles of microRNA single-nucleotide polymorphisms in cervical cancer.Int J Med Sci. 2021 Apr 7;18(11):2347-2354. doi: 10.7150/ijms.57990. eCollection 2021. Int J Med Sci. 2021. PMID: 33967611 Free PMC article. Review.
-
Long non-coding RNA NEAT1 contributes to lipopolysaccharide-induced inflammation and apoptosis of human middle ear epithelial cells via regulating the miR-301b-3p/TLR4 axis.Exp Ther Med. 2021 Dec;22(6):1360. doi: 10.3892/etm.2021.10795. Epub 2021 Sep 24. Exp Ther Med. 2021. PMID: 34659506 Free PMC article.
References
-
- Tsikouras P, Zervoudis S, Manav B, et al. Cervical cancer: screening, diagnosis and staging. J BUON. 2016;21:320–325. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

