Integrated nomogram based on five stage-related genes and TNM stage to predict 1-year recurrence in hepatocellular carcinoma
- PMID: 32368186
- PMCID: PMC7189530
- DOI: 10.1186/s12935-020-01216-9
Integrated nomogram based on five stage-related genes and TNM stage to predict 1-year recurrence in hepatocellular carcinoma
Abstract
Background: The primary tumor, regional lymph nodes and distant metastasis (TNM) stage is an independent risk factor for 1-year hepatocellular carcinoma (HCC) recurrence but has insufficient predictive efficiency. We attempt to develop and validate a nomogram to predict 1-year recurrence in HCC and improve the predictive efficiency of the TNM stage.
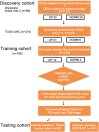
Methods: A total of 541 HCC patients were enrolled in the study. The risk score (RS) model was established with the logistic least absolute shrinkage and selector operation algorithm. The predictive nomogram was further validated in the internal testing cohort and external validation cohort. The area under the receiver operating characteristic curves (AUCs), decision curves and clinical impact curves were used to evaluate the predictive accuracy and clinical value of the nomogram.
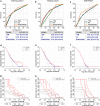
Results: In the training cohort, we identified a RS model consisting of five stage-related genes (NUP62, EHMT2, RANBP1, MSH6 and FHL2) for recurrence at 1 year. The 1-year disease-free survival of patients was worse in the high-risk group than in the low-risk group (P < 0.0001), and 1-year recurrence was more likely in the high-risk group (Hazard ratio: 3.199, P < 0.001). The AUC of the nomogram was 0.739, 0.718 and 0.693 in the training, testing and external validation cohort, respectively, and these values were larger than the corresponding AUC of the TNM stage (0.681, 0.688 and 0.616, respectively).
Conclusions: A RS model consisting of five stage-related genes was successfully identified for predicting 1-year HCC recurrence. Then, a novel nomogram based on the RS model and TNM stage to predict 1-year HCC recurrence was also developed and validated.
Keywords: 1-year recurrence; Hepatocellular carcinoma; Nomogram; Risk score model; TNM stage.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures



References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. - PubMed
-
- Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection. Ann Surg. 2015;261(5):947–955. - PubMed
-
- Poon RT. Differentiating early and late recurrences after resection of HCC in cirrhotic patients: implications on surveillance, prevention, and treatment strategies. Ann Surg Oncol. 2009;16(4):792–794. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

