MaRe: Processing Big Data with application containers on Apache Spark
- PMID: 32369166
- PMCID: PMC7199472
- DOI: 10.1093/gigascience/giaa042
MaRe: Processing Big Data with application containers on Apache Spark
Abstract
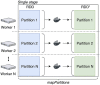
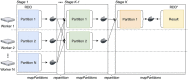
Background: Life science is increasingly driven by Big Data analytics, and the MapReduce programming model has been proven successful for data-intensive analyses. However, current MapReduce frameworks offer poor support for reusing existing processing tools in bioinformatics pipelines. Furthermore, these frameworks do not have native support for application containers, which are becoming popular in scientific data processing.
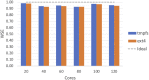
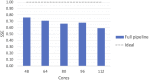
Results: Here we present MaRe, an open source programming library that introduces support for Docker containers in Apache Spark. Apache Spark and Docker are the MapReduce framework and container engine that have collected the largest open source community; thus, MaRe provides interoperability with the cutting-edge software ecosystem. We demonstrate MaRe on 2 data-intensive applications in life science, showing ease of use and scalability.
Conclusions: MaRe enables scalable data-intensive processing in life science with Apache Spark and application containers. When compared with current best practices, which involve the use of workflow systems, MaRe has the advantage of providing data locality, ingestion from heterogeneous storage systems, and interactive processing. MaRe is generally applicable and available as open source software.
Keywords: Apache Spark; Big Data; MapReduce; application containers; workflows.
© The Author(s) 2020. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

