The Regulation of Homologous Recombination by Helicases
- PMID: 32369918
- PMCID: PMC7290689
- DOI: 10.3390/genes11050498
The Regulation of Homologous Recombination by Helicases
Abstract
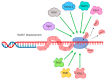
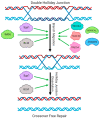
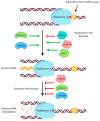
Homologous recombination is essential for DNA repair, replication and the exchange of genetic material between parental chromosomes during meiosis. The stages of recombination involve complex reorganization of DNA structures, and the successful completion of these steps is dependent on the activities of multiple helicase enzymes. Helicases of many different families coordinate the processing of broken DNA ends, and the subsequent formation and disassembly of the recombination intermediates that are necessary for template-based DNA repair. Loss of recombination-associated helicase activities can therefore lead to genomic instability, cell death and increased risk of tumor formation. The efficiency of recombination is also influenced by the 'anti-recombinase' effect of certain helicases, which can direct DNA breaks toward repair by other pathways. Other helicases regulate the crossover versus non-crossover outcomes of repair. The use of recombination is increased when replication forks and the transcription machinery collide, or encounter lesions in the DNA template. Successful completion of recombination in these situations is also regulated by helicases, allowing normal cell growth, and the maintenance of genomic integrity.
Keywords: DNA repair; anti-recombinase; helicase; recombination; replication; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Main Role of Srs2 in DNA Repair Depends on Its Helicase Activity, Rather than on Its Interactions with PCNA or Rad51.mBio. 2018 Jul 17;9(4):e01192-18. doi: 10.1128/mBio.01192-18. mBio. 2018. PMID: 30018112 Free PMC article.
-
Prevention of unwanted recombination at damaged replication forks.Curr Genet. 2020 Dec;66(6):1045-1051. doi: 10.1007/s00294-020-01095-7. Epub 2020 Jul 15. Curr Genet. 2020. PMID: 32671464 Free PMC article. Review.
-
DNA Damage Tolerance Pathway Choice Through Uls1 Modulation of Srs2 SUMOylation in Saccharomyces cerevisiae.Genetics. 2017 May;206(1):513-525. doi: 10.1534/genetics.116.196568. Epub 2017 Mar 24. Genetics. 2017. PMID: 28341648 Free PMC article.
-
Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing.PLoS Genet. 2013 Apr;9(4):e1003435. doi: 10.1371/journal.pgen.1003435. Epub 2013 Apr 11. PLoS Genet. 2013. PMID: 23593030 Free PMC article.
-
Limiting homologous recombination at stalled replication forks is essential for cell viability: DNA2 to the rescue.Curr Genet. 2020 Dec;66(6):1085-1092. doi: 10.1007/s00294-020-01106-7. Epub 2020 Sep 9. Curr Genet. 2020. PMID: 32909097 Free PMC article. Review.
Cited by
-
Neural Tube Defects and Folate Deficiency: Is DNA Repair Defective?Int J Mol Sci. 2023 Jan 22;24(3):2220. doi: 10.3390/ijms24032220. Int J Mol Sci. 2023. PMID: 36768542 Free PMC article. Review.
-
Motifs of the C-terminal domain of MCM9 direct localization to sites of mitomycin-C damage for RAD51 recruitment.J Biol Chem. 2021 Jan-Jun;296:100355. doi: 10.1016/j.jbc.2021.100355. Epub 2021 Feb 2. J Biol Chem. 2021. PMID: 33539926 Free PMC article.
-
The toposiomerase IIIalpha-RMI1-RMI2 complex orients human Bloom's syndrome helicase for efficient disruption of D-loops.Nat Commun. 2022 Feb 3;13(1):654. doi: 10.1038/s41467-022-28208-9. Nat Commun. 2022. PMID: 35115525 Free PMC article.
-
Homologous Recombination Subpathways: A Tangle to Resolve.Front Genet. 2021 Aug 2;12:723847. doi: 10.3389/fgene.2021.723847. eCollection 2021. Front Genet. 2021. PMID: 34408777 Free PMC article. Review.
-
Fission yeast Rad54 prevents intergenerational buildup of Rad51 aggregates in proliferating cells.Life Sci Alliance. 2025 Aug 18;8(11):e202503252. doi: 10.26508/lsa.202503252. Print 2025 Nov. Life Sci Alliance. 2025. PMID: 40825586 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

