Deciphering the Role of Colicins during Colonization of the Mammalian Gut by Commensal E. coli
- PMID: 32370119
- PMCID: PMC7284606
- DOI: 10.3390/microorganisms8050664
Deciphering the Role of Colicins during Colonization of the Mammalian Gut by Commensal E. coli
Abstract
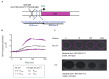
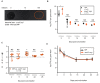
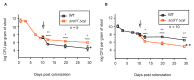
Colicins are specific and potent toxins produced by Enterobacteriaceae that result in the rapid elimination of sensitive cells. Colicin production is commonly found throughout microbial populations, suggesting its potential importance for bacterial survival in complex microbial environments. Nonetheless, as colicin biology has been predominately studied using synthetic models, it remains unclear how colicin production contributes to survival and fitness of a colicin-producing commensal strain in a natural environment. To address this gap, we took advantage of MP1, an E. coli strain that harbors a colicinogenic plasmid and is a natural colonizer of the murine gut. Using this model, we validated that MP1 is competent for colicin production and then directly interrogated the importance of colicin production and immunity for MP1 survival in the murine gut. We showed that colicin production is dispensable for sustained colonization in the unperturbed gut. A strain lacking colicin production or immunity shows minimal fitness defects and can resist displacement by colicin producers. This report extends our understanding of the role that colicin production may play for E. coli during gut colonization and suggests that colicin production is not essential for a commensal to persist in its physiologic niche in the absence of exogenous challenges.
Keywords: DNA damage response; colicin; colonization; commensal E. coli.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Haag L.M., Fischer A., Otto B., Plickert R., Kühl A.A., Göbel U.B., Bereswill S., Heimesaat M.M. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS ONE. 2012;7:e35988. doi: 10.1371/journal.pone.0035988. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

