Molecular Signatures of JMJD10/ MINA53 in Gastric Cancer
- PMID: 32370161
- PMCID: PMC7281541
- DOI: 10.3390/cancers12051141
Molecular Signatures of JMJD10/ MINA53 in Gastric Cancer
Abstract
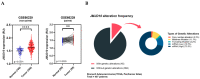
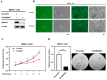
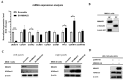
The JMJD10 gene and its encoded protein MYC-induced nuclear antigen (MINA53) are associated with multiple cancers. Besides having both an oncogenic and tumor suppressor function, the intricate role of JMJD10 in cancer is complex as it depends on the cancer type. In particular, the functional role of JMJD10/MINA53 in gastric cancer has been poorly understood. In this study, we have unraveled the molecular signatures and functional roles of JMJD10/MINA53 in gastric cancer by multiple approaches, i.e., multi-omics bioinformatics study, analysis of human gastric cancer tissues, and studies in vitro using knockdown or overexpression strategies in gastric cancer cell lines. The results indicated that the JMJD10 gene and MINA53 protein are commonly overexpressed in cancer patients. JMJD10/MINA53 is involved in the regulation of proliferation and survival of gastric cancer by controlling cell cycle gene expression. These processes are highly associated with MINA53 enzymatic activity in the regulation of H3K9me3 methylation status and controlling activation of AP-1 signaling pathways. This highlights the oncogenic role of JMJD10/MINA53 in gastric cancer and opens the opportunity to develop therapeutic targeting of JMJD10/MINA53 in gastric cancer.
Keywords: JMJD10/MINA53; KDM; gastric cancer; histone demethylase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

