An Unsupervised Strategy for Identifying Epithelial-Mesenchymal Transition State Metrics in Breast Cancer and Melanoma
- PMID: 32371374
- PMCID: PMC7200934
- DOI: 10.1016/j.isci.2020.101080
An Unsupervised Strategy for Identifying Epithelial-Mesenchymal Transition State Metrics in Breast Cancer and Melanoma
Abstract
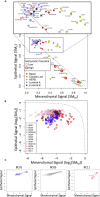
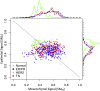
Digital cytometry aims to identify different cell types in the tumor microenvironment, with the current focus on immune cells. Yet, identifying how changes in tumor cell phenotype, such as the epithelial-mesenchymal transition, influence the immune contexture is emerging as an important question. To extend digital cytometry, we developed an unsupervised feature extraction and selection strategy to capture functional plasticity tailored to breast cancer and melanoma separately. Specifically, principal component analysis coupled with resampling helped develop gene expression-based state metrics that characterize differentiation within an epithelial to mesenchymal-like state space and independently correlate with metastatic potential. First developed using cell lines, the orthogonal state metrics were refined to exclude the contributions of normal fibroblasts and provide tissue-level state estimates using bulk tissue RNA-seq measures. The resulting metrics for differentiation state aim to inform a more holistic view of how the malignant cell phenotype influences the immune contexture within the tumor microenvironment.
Keywords: Bioinformatics; Cancer; Stem Cells Research.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing financial interests.
Figures









References
-
- Alon U. volume 10. Chapman & Hall/CRC; 2007. An introduction to systems biology: design principles of biological circuits; pp. 97–104. (Chapman & Hall/CRC Mathematical and Computational Biology Series).
-
- Alonso S.R., Tracey L., Ortiz P., Perez-Gomez B., Palacios J., Pollan M., Linares J., Serrano S., Saez-Castillo A.I., Sanchez L. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. - PubMed
-
- American Cancer Society . American Cancer Society; 2019. Cancer Facts & Figures 2019.
Grants and funding
LinkOut - more resources
Full Text Sources

