RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers
- PMID: 32371455
- PMCID: PMC7373991
- DOI: 10.1261/rna.075341.120
RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers
Abstract
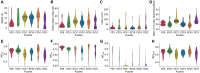
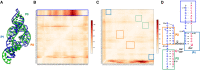
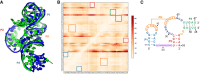
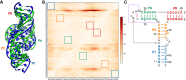
RNA-Puzzles is a collective endeavor dedicated to the advancement and improvement of RNA 3D structure prediction. With agreement from crystallographers, the RNA structures are predicted by various groups before the publication of the crystal structures. We now report the prediction of 3D structures for six RNA sequences: four nucleolytic ribozymes and two riboswitches. Systematic protocols for comparing models and crystal structures are described and analyzed. In these six puzzles, we discuss (i) the comparison between the automated web servers and human experts; (ii) the prediction of coaxial stacking; (iii) the prediction of structural details and ligand binding; (iv) the development of novel prediction methods; and (v) the potential improvements to be made. We show that correct prediction of coaxial stacking and tertiary contacts is essential for the prediction of RNA architecture, while ligand binding modes can only be predicted with low resolution and simultaneous prediction of RNA structure with accurate ligand binding still remains out of reach. All the predicted models are available for the future development of force field parameters and the improvement of comparison and assessment tools.
Keywords: RNA structure; aptamer; modeling; prediction; ribozyme.
© 2020 Miao et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM134919/GM/NIGMS NIH HHS/United States
- R01 GM114015/GM/NIGMS NIH HHS/United States
- R01 GM117059/GM/NIGMS NIH HHS/United States
- R01 GM073850/GM/NIGMS NIH HHS/United States
- R01 GM063732/GM/NIGMS NIH HHS/United States
- R35 GM134864/GM/NIGMS NIH HHS/United States
- UL1 TR002014/TR/NCATS NIH HHS/United States
- R01 GM123247/GM/NIGMS NIH HHS/United States
- R21 CA219847/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R35 GM122579/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R21 GM102716/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
