Isolation and Characterization of the Novel Phage JD032 and Global Transcriptomic Response during JD032 Infection of Clostridioides difficile Ribotype 078
- PMID: 32371470
- PMCID: PMC7205517
- DOI: 10.1128/mSystems.00017-20
Isolation and Characterization of the Novel Phage JD032 and Global Transcriptomic Response during JD032 Infection of Clostridioides difficile Ribotype 078
Abstract
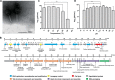
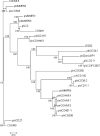
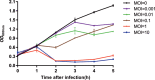
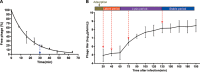
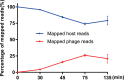
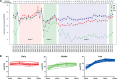
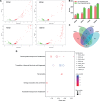
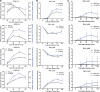
Insights into the interaction between phages and their bacterial hosts are crucial for the development of phage therapy. However, only one study has investigated global gene expression of Clostridioides (formerly Clostridium) difficile carrying prophage, and transcriptional reprogramming during lytic infection has not been studied. Here, we presented the isolation, propagation, and characterization of a newly discovered 35,109-bp phage, JD032, and investigated the global transcriptomes of both JD032 and C. difficile ribotype 078 (RT078) strain TW11 during JD032 infection. Transcriptome sequencing (RNA-seq) revealed the progressive replacement of bacterial host mRNA with phage transcripts. The expressed genes of JD032 were clustered into early, middle, and late temporal categories that were functionally similar. Specifically, a gene (JD032_orf016) involved in the lysis-lysogeny decision was identified as an early expression gene. Only 17.7% (668/3,781) of the host genes were differentially expressed, and more genes were downregulated than upregulated. The expression of genes involved in host macromolecular synthesis (DNA/RNA/proteins) was altered by JD032 at the level of transcription. In particular, the expression of the ropA operon was downregulated. Most noteworthy is that the gene expression of some antiphage systems, including CRISPR-Cas, restriction-modification, and toxin-antitoxin systems, was suppressed by JD032 during infection. In addition, bacterial sporulation, adhesion, and virulence factor genes were significantly downregulated. This study provides the first description of the interaction between anaerobic spore-forming bacteria and phages during lytic infection and highlights new aspects of C. difficile phage-host interactions.IMPORTANCE C. difficile is one of the most clinically significant intestinal pathogens. Although phages have been shown to effectively control C. difficile infection, the host responses to phage predation have not been fully studied. In this study, we reported the isolation and characterization of a new phage, JD032, and analyzed the global transcriptomic changes in the hypervirulent RT078 C. difficile strain, TW11, during phage JD032 infection. We found that bacterial host mRNA was progressively replaced with phage transcripts, three temporal categories of JD032 gene expression, the extensive interplay between phage-bacterium, antiphage-like responses of the host and phage evasion, and decreased expression of sporulation- and virulence-related genes of the host after phage infection. These findings confirmed the complexity of interactions between C. difficile and phages and suggest that phages undergoing a lytic cycle may also cause different phenotypes in hosts, similar to prophages, which may inspire phage therapy for the control of C. difficile.
Keywords: Clostridioides difficile; RNA-seq; bacteria-phage interaction; bacteriophage; ribotype 078; transcriptome.
Copyright © 2020 Li et al.
Figures
References
-
- Walker AS, Eyre DW, Wyllie DH, Dingle KE, Griffiths D, Shine B, Oakley S, O’Connor L, Finney J, Vaughan A, Crook DW, Wilcox MH, Peto TEA, Infections in Oxfordshire Research Database (IORD). 2013. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin Infect Dis 56:1589–1600. doi: 10.1093/cid/cit127. - DOI - PMC - PubMed
-
- Knetsch CW, Kumar N, Forster SC, Connor TR, Browne HP, Harmanus C, Sanders IM, Harris SR, Turner L, Morris T, Perry M, Miyajima F, Roberts P, Pirmohamed M, Songer JG, Weese JS, Indra A, Corver J, Rupnik M, Wren BW, Riley TV, Kuijper EJ, Lawley TD. 2017. Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans. J Clin Microbiol 56:e01384-17. doi: 10.1128/JCM.01384-17. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

