Metabolic cost of rapid adaptation of single yeast cells
- PMID: 32371488
- PMCID: PMC7245094
- DOI: 10.1073/pnas.1913767117
Metabolic cost of rapid adaptation of single yeast cells
Abstract
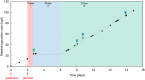
Cells can rapidly adapt to changing environments through nongenetic processes; however, the metabolic cost of such adaptation has never been considered. Here we demonstrate metabolic coupling in a remarkable, rapid adaptation process (1 in 1,000 cells adapt per hour) by simultaneously measuring metabolism and division of thousands of individual Saccharomyces cerevisiae cells using a droplet microfluidic system: droplets containing single cells are immobilized in a two-dimensional (2D) array, with osmotically induced changes in droplet volume being used to measure cell metabolism, while simultaneously imaging the cells to measure division. Following a severe challenge, most cells, while not dividing, continue to metabolize, displaying a remarkably wide diversity of metabolic trajectories from which adaptation events can be anticipated. Adaptation requires a characteristic amount of energy, indicating that it is an active process. The demonstration that metabolic trajectories predict a priori adaptation events provides evidence of tight energetic coupling between metabolism and regulatory reorganization in adaptation. This process allows S. cerevisiae to adapt on a physiological timescale, but related phenomena may also be important in other processes, such as cellular differentiation, cellular reprogramming, and the emergence of drug resistance in cancer.
Keywords: adaptation; droplet-based microfluidics; genetic rewiring; single-cell metabolism.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Robinson B. W., Dukas R., The influence of phenotypic modifications on evolution: The Baldwin effect and modern perspectives. Oikos 85, 582–589 (1999).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

