Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins
- PMID: 32371491
- PMCID: PMC7245119
- DOI: 10.1073/pnas.1920097117
Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins
Abstract
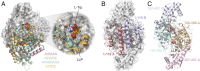
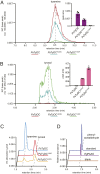
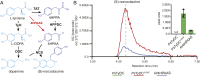
Radiation of the plant pyridoxal 5'-phosphate (PLP)-dependent aromatic l-amino acid decarboxylase (AAAD) family has yielded an array of paralogous enzymes exhibiting divergent substrate preferences and catalytic mechanisms. Plant AAADs catalyze either the decarboxylation or decarboxylation-dependent oxidative deamination of aromatic l-amino acids to produce aromatic monoamines or aromatic acetaldehydes, respectively. These compounds serve as key precursors for the biosynthesis of several important classes of plant natural products, including indole alkaloids, benzylisoquinoline alkaloids, hydroxycinnamic acid amides, phenylacetaldehyde-derived floral volatiles, and tyrosol derivatives. Here, we present the crystal structures of four functionally distinct plant AAAD paralogs. Through structural and functional analyses, we identify variable structural features of the substrate-binding pocket that underlie the divergent evolution of substrate selectivity toward indole, phenyl, or hydroxyphenyl amino acids in plant AAADs. Moreover, we describe two mechanistic classes of independently arising mutations in AAAD paralogs leading to the convergent evolution of the derived aldehyde synthase activity. Applying knowledge learned from this study, we successfully engineered a shortened benzylisoquinoline alkaloid pathway to produce (S)-norcoclaurine in yeast. This work highlights the pliability of the AAAD fold that allows change of substrate selectivity and access to alternative catalytic mechanisms with only a few mutations.
Keywords: AAAD; aromatic amino acid metabolism; enzyme evolution; specialized metabolism.
Conflict of interest statement
Competing interest statement: J.-K.W. is a cofounder, a member of the Scientific Advisory Board, and a shareholder of DoubleRainbow Biosciences, which develops biotechnologies related to natural products.
Figures


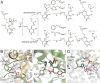
Similar articles
-
Aromatic L-amino acid decarboxylases: mechanistic features and microbial applications.Appl Microbiol Biotechnol. 2022 Jun;106(12):4445-4458. doi: 10.1007/s00253-022-12028-4. Epub 2022 Jun 28. Appl Microbiol Biotechnol. 2022. PMID: 35763068 Review.
-
Active-Site Oxygen Accessibility and Catalytic Loop Dynamics of Plant Aromatic Amino Acid Decarboxylases from Molecular Simulations.Biochemistry. 2024 Aug 6;63(15):1980-1990. doi: 10.1021/acs.biochem.4c00144. Epub 2024 Jul 15. Biochemistry. 2024. PMID: 39008055 Free PMC article.
-
Biochemical evaluation of the decarboxylation and decarboxylation-deamination activities of plant aromatic amino acid decarboxylases.J Biol Chem. 2013 Jan 25;288(4):2376-87. doi: 10.1074/jbc.M112.401752. Epub 2012 Nov 30. J Biol Chem. 2013. PMID: 23204519 Free PMC article.
-
Diverse functional evolution of serine decarboxylases: identification of two novel acetaldehyde synthases that uses hydrophobic amino acids as substrates.BMC Plant Biol. 2014 Sep 18;14:247. doi: 10.1186/s12870-014-0247-x. BMC Plant Biol. 2014. PMID: 25230835 Free PMC article.
-
Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications.Phytochemistry. 2000 May;54(2):121-38. doi: 10.1016/s0031-9422(00)00050-9. Phytochemistry. 2000. PMID: 10872203 Review.
Cited by
-
Evolution of plant metabolism: the state-of-the-art.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230347. doi: 10.1098/rstb.2023.0347. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343029 Free PMC article. Review.
-
Aromatic L-amino acid decarboxylases: mechanistic features and microbial applications.Appl Microbiol Biotechnol. 2022 Jun;106(12):4445-4458. doi: 10.1007/s00253-022-12028-4. Epub 2022 Jun 28. Appl Microbiol Biotechnol. 2022. PMID: 35763068 Review.
-
A Collaborative Classroom Investigation of the Evolution of SABATH Methyltransferase Substrate Preference Shifts over 120 My of Flowering Plant History.Mol Biol Evol. 2022 Mar 2;39(3):msac007. doi: 10.1093/molbev/msac007. Mol Biol Evol. 2022. PMID: 35021222 Free PMC article.
-
Transcriptomics and network pharmacology analysis reveal key genes in alkaloid biosynthesis in Zephyranthes candida and therapeutic targets for LIHC.Planta. 2025 Jul 10;262(2):45. doi: 10.1007/s00425-025-04756-4. Planta. 2025. PMID: 40640563
-
In Vitro Psilocybin Synthesis by Co-Immobilized Enzymes.Chemistry. 2025 May 22;31(29):e202501037. doi: 10.1002/chem.202501037. Epub 2025 Apr 21. Chemistry. 2025. PMID: 40202903 Free PMC article.
References
-
- Weng J.-K., Philippe R. N., Noel J. P., The rise of chemodiversity in plants. Science 336, 1667–1670 (2012). - PubMed
-
- Chae L., Kim T., Nilo-Poyanco R., Rhee S. Y., Genomic signatures of specialized metabolism in plants. Science 344, 510–513 (2014). - PubMed
-
- Kaltenbach M. et al. ., Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol. 14, 548–555 (2018). - PubMed
-
- Burkhard P., Dominici P., Borri-Voltattorni C., Jansonius J. N., Malashkevich V. N., Structural insight into Parkinson’s disease treatment from drug-inhibited DOPA decarboxylase. Nat. Struct. Biol. 8, 963–967 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

