The calcitonin-like system is an ancient regulatory system of biomineralization
- PMID: 32371888
- PMCID: PMC7200681
- DOI: 10.1038/s41598-020-64118-w
The calcitonin-like system is an ancient regulatory system of biomineralization
Abstract
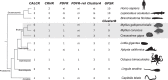
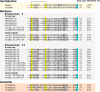
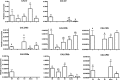
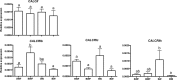
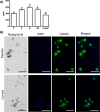
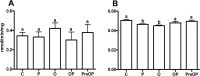
Biomineralization is the process by which living organisms acquired the capacity to accumulate minerals in tissues. Shells are the biomineralized exoskeleton of marine molluscs produced by the mantle but factors that regulate mantle shell building are still enigmatic. This study sought to identify candidate regulatory factors of molluscan shell mineralization and targeted family B G-protein coupled receptors (GPCRs) and ligands that include calcium regulatory factors in vertebrates, such as calcitonin (CALC). In molluscs, CALC receptor (CALCR) number was variable and arose through lineage and species-specific duplications. The Mediterranean mussel (Mytilus galloprovincialis) mantle transcriptome expresses six CALCR-like and two CALC-precursors encoding four putative mature peptides. Mussel CALCR-like are activated in vitro by vertebrate CALC but only receptor CALCRIIc is activated by the mussel CALCIIa peptide (EC50 = 2.6 ×10-5 M). Ex-vivo incubations of mantle edge tissue and mantle cells with CALCIIa revealed they accumulated significantly more calcium than untreated tissue and cells. Mussel CALCIIa also significantly decreased mantle acid phosphatase activity, which is associated with shell remodelling. Our data indicate the CALC-like system as candidate regulatory factors of shell mineralization. The identification of the CALC system from molluscs to vertebrates suggests it is an ancient and conserved calcium regulatory system of mineralization.
Conflict of interest statement
The authors declare no competing interests.
Figures

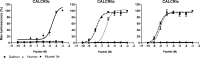
Similar articles
-
Neuropeptides regulate shell growth in the Mediterranean mussel (Mytilus galloprovincialis).Int J Biol Macromol. 2024 Nov;281(Pt 4):136500. doi: 10.1016/j.ijbiomac.2024.136500. Epub 2024 Oct 15. Int J Biol Macromol. 2024. PMID: 39419152
-
Allatostatin-type A, kisspeptin and galanin GPCRs and putative ligands as candidate regulatory factors of mantle function.Mar Genomics. 2016 Jun;27:25-35. doi: 10.1016/j.margen.2015.12.003. Epub 2015 Dec 30. Mar Genomics. 2016. PMID: 26751715 Review.
-
Core genes of biomineralization and cis-regulatory long non-coding RNA regulate shell growth in bivalves.J Adv Res. 2024 Oct;64:117-129. doi: 10.1016/j.jare.2023.11.024. Epub 2023 Nov 22. J Adv Res. 2024. PMID: 37995944 Free PMC article.
-
Mantle transcriptome sequencing of Mytilus spp. and identification of putative biomineralization genes.PeerJ. 2019 Jan 14;6:e6245. doi: 10.7717/peerj.6245. eCollection 2019. PeerJ. 2019. PMID: 30723611 Free PMC article.
-
Sea shell diversity and rapidly evolving secretomes: insights into the evolution of biomineralization.Front Zool. 2016 Jun 7;13:23. doi: 10.1186/s12983-016-0155-z. eCollection 2016. Front Zool. 2016. PMID: 27279892 Free PMC article. Review.
Cited by
-
Molecular mechanisms of biomineralization in marine invertebrates.J Exp Biol. 2020 May 29;223(Pt 11):jeb206961. doi: 10.1242/jeb.206961. J Exp Biol. 2020. PMID: 32471885 Free PMC article. Review.
-
Association analysis of production traits of Japanese quail (Coturnix japonica) using restriction-site associated DNA sequencing.Sci Rep. 2023 Dec 2;13(1):21307. doi: 10.1038/s41598-023-48293-0. Sci Rep. 2023. PMID: 38042890 Free PMC article.
-
Nemertean, Brachiopod, and Phoronid Neuropeptidomics Reveals Ancestral Spiralian Signaling Systems.Mol Biol Evol. 2021 Oct 27;38(11):4847-4866. doi: 10.1093/molbev/msab211. Mol Biol Evol. 2021. PMID: 34272863 Free PMC article.
-
Genome-Wide Association Study-Based Identification of SNPs and Haplotypes Associated With Goose Reproductive Performance and Egg Quality.Front Genet. 2021 Mar 12;12:602583. doi: 10.3389/fgene.2021.602583. eCollection 2021. Front Genet. 2021. PMID: 33777090 Free PMC article.
-
Pituitary whole transcriptome analysis reveals key genes regulating reproduction in Hy-Line Brown hens and the construction of their ceRNA molecular regulatory network.BMC Genomics. 2024 Nov 18;25(1):1100. doi: 10.1186/s12864-024-11035-1. BMC Genomics. 2024. PMID: 39558278 Free PMC article.
References
-
- Suzuki M, Nagasawa H. Mollusk shell structures and their formation mechanism. Can. J. Zool. Can. Zool. 2013;91:349–366. doi: 10.1139/cjz-2012-0333. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

