Three-dimensional nanoscopy of whole cells and tissues with in situ point spread function retrieval
- PMID: 32371980
- PMCID: PMC7289454
- DOI: 10.1038/s41592-020-0816-x
Three-dimensional nanoscopy of whole cells and tissues with in situ point spread function retrieval
Abstract
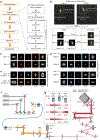
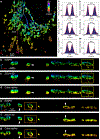
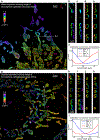
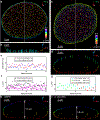
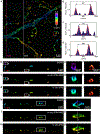
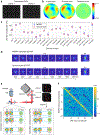
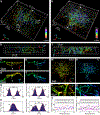
Single-molecule localization microscopy is a powerful tool for visualizing subcellular structures, interactions and protein functions in biological research. However, inhomogeneous refractive indices inside cells and tissues distort the fluorescent signal emitted from single-molecule probes, which rapidly degrades resolution with increasing depth. We propose a method that enables the construction of an in situ 3D response of single emitters directly from single-molecule blinking datasets, and therefore allows their locations to be pinpointed with precision that achieves the Cramér-Rao lower bound and uncompromised fidelity. We demonstrate this method, named in situ PSF retrieval (INSPR), across a range of cellular and tissue architectures, from mitochondrial networks and nuclear pores in mammalian cells to amyloid-β plaques and dendrites in brain tissues and elastic fibers in developing cartilage of mice. This advancement expands the routine applicability of super-resolution microscopy from selected cellular targets near coverslips to intra- and extracellular targets deep inside tissues.
Conflict of interest statement
Competing interests
F.X., D.M., and F.H. are inventors on patent application submitted by Purdue University that covers basic principles of INSPR.
Figures









References
-
- Hell SW & Wichmann J Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett 19, 780–782 (1994). - PubMed
-
- Wu Y & Shroff H Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat. Methods 15, 1011–1019 (2018). - PubMed
-
- Betzig E et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

