Phylotastic: Improving Access to Tree-of-Life Knowledge With Flexible, on-the-Fly Delivery of Trees
- PMID: 32372858
- PMCID: PMC7192527
- DOI: 10.1177/1176934319899384
Phylotastic: Improving Access to Tree-of-Life Knowledge With Flexible, on-the-Fly Delivery of Trees
Abstract
A comprehensive phylogeny of species, i.e., a tree of life, has potential uses in a variety of contexts, including research, education, and public policy. Yet, accessing the tree of life typically requires special knowledge, complex software, or long periods of training. The Phylotastic project aims make it as easy to get a phylogeny of species as it is to get driving directions from mapping software. In prior work, we presented a design for an open system to validate and manage taxon names, find phylogeny resources, extract subtrees matching a user's taxon list, scale trees to time, and integrate related resources such as species images. Here, we report the implementation of a set of tools that together represent a robust, accessible system for on-the-fly delivery of phylogenetic knowledge. This set of tools includes a web portal to execute several customizable workflows to obtain species phylogenies (scaled by geologic time and decorated with thumbnail images); more than 30 underlying web services (accessible via a common registry); and code toolkits in R and Python (allowing others to develop custom applications using Phylotastic services). The Phylotastic system, accessible via http://www.phylotastic.org, provides a unique resource to access the current state of phylogenetic knowledge, useful for a variety of cases in which a tree extracted quickly from online resources (as distinct from a tree custom-made from character data) is sufficient, as it is for many casual uses of trees identified here.
Keywords: Phylogeny; tree of life; web services.
© The Author(s) 2020.
Conflict of interest statement
Declaration of conflicting interest:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
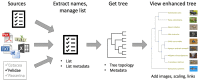
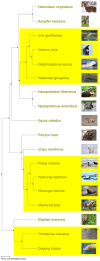
Update of
- doi: 10.1101/419143
Similar articles
-
Phylotastic! Making tree-of-life knowledge accessible, reusable and convenient.BMC Bioinformatics. 2013 May 13;14:158. doi: 10.1186/1471-2105-14-158. BMC Bioinformatics. 2013. PMID: 23668630 Free PMC article.
-
DateLife: Leveraging Databases and Analytical Tools to Reveal the Dated Tree of Life.Syst Biol. 2024 Jul 27;73(2):470-485. doi: 10.1093/sysbio/syae015. Syst Biol. 2024. PMID: 38507308 Free PMC article.
-
PhyloExplorer: a web server to validate, explore and query phylogenetic trees.BMC Evol Biol. 2009 May 18;9:108. doi: 10.1186/1471-2148-9-108. BMC Evol Biol. 2009. PMID: 19450253 Free PMC article.
-
A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway.Evol Bioinform Online. 2015 Mar 16;11:43-8. doi: 10.4137/EBO.S21501. eCollection 2015. Evol Bioinform Online. 2015. PMID: 25861210 Free PMC article. Review.
-
Insect phylogenetics in the digital age.Curr Opin Insect Sci. 2016 Dec;18:48-52. doi: 10.1016/j.cois.2016.09.008. Epub 2016 Oct 18. Curr Opin Insect Sci. 2016. PMID: 27939710 Review.
References
-
- Cracraft J, Donoghue M, Dragoo J, Hillis D, Yates T. Assembling the tree of life: harnessing life’s history to benefit science and society. http://ucjeps.berkeley.edu/tol.pdf. Report, University of California, Berkeley: Published 2002. Accessed December 24, 2019.
Publication types
LinkOut - more resources
Full Text Sources

