LysM Proteins Regulate Fungal Development and Contribute to Hyphal Protection and Biocontrol Traits in Clonostachys rosea
- PMID: 32373095
- PMCID: PMC7176902
- DOI: 10.3389/fmicb.2020.00679
LysM Proteins Regulate Fungal Development and Contribute to Hyphal Protection and Biocontrol Traits in Clonostachys rosea
Abstract
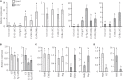
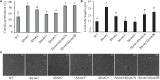
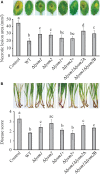
Lysin motif (LysM) modules are approximately 50 amino acids long and bind to peptidoglycan, chitin and its derivatives. Certain LysM proteins in plant pathogenic and entomopathogenic fungi are shown to scavenge chitin oligosaccharides and thereby dampen host defense reactions. Other LysM proteins can protect the fungal cell wall against hydrolytic enzymes. In this study, we investigated the biological function of LysM proteins in the mycoparasitic fungus Clonostachys rosea. The C. rosea genome contained three genes coding for LysM-containing proteins and gene expression analysis revealed that lysm1 and lysm2 were induced during mycoparasitic interaction with Fusarium graminearum and during colonization of wheat roots. Lysm1 was suppressed in germinating conidia, while lysm2 was induced during growth in chitin or peptidoglycan-containing medium. Deletion of lysm1 and lysm2 resulted in mutants with increased levels of conidiation and conidial germination, but reduced ability to control plant diseases caused by F. graminearum and Botrytis cinerea. The Δlysm2 strain showed a distinct, accelerated mycelial disintegration phenotype accompanied by reduced biomass production and hyphal protection against hydrolytic enzymes including chitinases, suggesting a role of LYSM2 in hyphal protection against chitinases. The Δlysm2 and Δlysm1Δlysm2 strains displayed reduced ability to colonize wheat roots, while only Δlysm1Δlysm2 failed to suppress expression of the wheat defense response genes PR1 and PR4. Based on our data, we propose a role of LYSM1 as a regulator of fungal development and of LYSM2 in cell wall protection against endogenous hydrolytic enzymes, while both are required to suppress plant defense responses. Our findings expand the understanding of the role of LysM proteins in fungal-fungal interactions and biocontrol.
Keywords: Clonostachys rosea; LysM effector; LysM protein; antagonism; biocontrol; biological control; fungal–fungal interactions; mycoparasitism.
Copyright © 2020 Dubey, Vélëz, Broberg, Jensen and Karlsson.
Figures





References
LinkOut - more resources
Full Text Sources

