A Primer for Microbiome Time-Series Analysis
- PMID: 32373155
- PMCID: PMC7186479
- DOI: 10.3389/fgene.2020.00310
A Primer for Microbiome Time-Series Analysis
Abstract
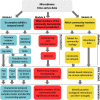
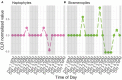
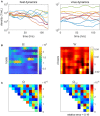
Time-series can provide critical insights into the structure and function of microbial communities. The analysis of temporal data warrants statistical considerations, distinct from comparative microbiome studies, to address ecological questions. This primer identifies unique challenges and approaches for analyzing microbiome time-series. In doing so, we focus on (1) identifying compositionally similar samples, (2) inferring putative interactions among populations, and (3) detecting periodic signals. We connect theory, code and data via a series of hands-on modules with a motivating biological question centered on marine microbial ecology. The topics of the modules include characterizing shifts in community structure and activity, identifying expression levels with a diel periodic signal, and identifying putative interactions within a complex community. Modules are presented as self-contained, open-access, interactive tutorials in R and Matlab. Throughout, we highlight statistical considerations for dealing with autocorrelated and compositional data, with an eye to improving the robustness of inferences from microbiome time-series. In doing so, we hope that this primer helps to broaden the use of time-series analytic methods within the microbial ecology research community.
Keywords: clustering; code:R; code:matlab; inference; marine microbiology; microbial ecology; periodicity; time-series analysis.
Copyright © 2020 Coenen, Hu, Luo, Muratore and Weitz.
Figures



References
-
- Agrawal A., Verschueren R., Diamond S., Boyd S. (2018). A rewriting system for convex optimization problems. J. Control Decis. 5, 42–60. 10.1080/23307706.2017.1397554 - DOI
-
- Aitchison J. (1983). The statistical analysis of compositional data. J. Int. Assoc. Math. Geol. 44, 139–177.
-
- Aitchison J. A., Vidal C., Martín-Fernández J., Pawlowsky-Glahn V. (2000). Logratio analysis and compositional distance. Math. Geol. 32, 271–275. 10.1023/A:1007529726302 - DOI
LinkOut - more resources
Full Text Sources

