PTPROt aggravates inflammation by enhancing NF-κB activation in liver macrophages during nonalcoholic steatohepatitis
- PMID: 32373213
- PMCID: PMC7196286
- DOI: 10.7150/thno.42658
PTPROt aggravates inflammation by enhancing NF-κB activation in liver macrophages during nonalcoholic steatohepatitis
Erratum in
-
Erratum: PTPROt aggravates inflammation by enhancing NF-κB activation in liver macrophages during nonalcoholic steatohepatitis: Erratum.Theranostics. 2022 Apr 26;12(8):3604-3606. doi: 10.7150/thno.73372. eCollection 2022. Theranostics. 2022. PMID: 35664079 Free PMC article.
Abstract
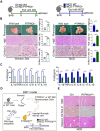
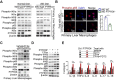
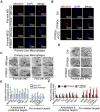
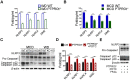
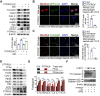
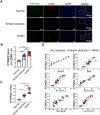
Rationale: Inflammation plays a crucial role in the progression of nonalcoholic steatohepatitis (NASH). Protein tyrosine phosphatase receptor type O truncated isoform (PTPROt) is an integral membrane protein that has been identified in osteoclasts, macrophages, and B lymphocytes. However, its relationship between inflammation and NASH is largely unknown. Herein, we aimed to study the function of PTPROt in NASH progression. Methods: We established a NASH mouse model in wild-type (WT), PTPRO knockout mice by western diet (WD) and methionine-choline-deficient diet (MCD). In addition, MCD-induced NASH model was established in BMT mice. Moreover, we determined the expression of PTPROt in liver macrophages in human subjects without steatosis, with simple steatosis, and with NASH to confirm the relationship between PTPROt and NASH. In vitro assays were also performed to study the molecular role of PTPROt in NASH progression. Results: Human samples and animal model results illustrated that PTPROt is increased in liver macrophages during NASH progression and is positively correlated with the degree of NASH. Our animal model also showed that PTPROt in liver macrophages can enhance the activation of the NF-κB signaling pathway, which induces the transcription of genes involved in the inflammatory response. Moreover, PTPROt promotes the transcription of pro-oxidant genes and inhibits antioxidant and protective genes via increased activation of the NF-κB signaling pathway, thereby causing an increased level of reactive oxygen species (ROS) and damaged mitochondria. This triggers the NLRP3-IL1β axis and causes a heightened inflammatory response. Notably, PTPROt partially limits inflammation and ROS production by promoting mitophagy, which participates in a negative feedback loop in this model. Conclusions: Our data strongly indicate that PTPROt plays a dual role in inflammation via the NF-κB signaling pathway in liver macrophages during NASH. Further studies are required to explore therapeutic strategies and prevention of this common liver disease through PTPROt.
Keywords: Inflammasome; Mitochondria; Mitophagy; NLR family pyrin domain containing 3; Reactive oxygen species.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
Fibrinogen-like protein 2 aggravates nonalcoholic steatohepatitis via interaction with TLR4, eliciting inflammation in macrophages and inducing hepatic lipid metabolism disorder.Theranostics. 2020 Aug 1;10(21):9702-9720. doi: 10.7150/thno.44297. eCollection 2020. Theranostics. 2020. PMID: 32863955 Free PMC article.
-
SOCS2 Suppresses Inflammation and Apoptosis during NASH Progression through Limiting NF-κB Activation in Macrophages.Int J Biol Sci. 2021 Oct 11;17(15):4165-4175. doi: 10.7150/ijbs.63889. eCollection 2021. Int J Biol Sci. 2021. PMID: 34803490 Free PMC article.
-
BMP9 promotes methionine- and choline-deficient diet-induced nonalcoholic steatohepatitis in non-obese mice by enhancing NF-κB dependent macrophage polarization.Int Immunopharmacol. 2021 Jul;96:107591. doi: 10.1016/j.intimp.2021.107591. Epub 2021 Mar 31. Int Immunopharmacol. 2021. PMID: 33812253
-
Tumor necrosis factor-α signaling in nonalcoholic steatohepatitis and targeted therapies.J Genet Genomics. 2022 Apr;49(4):269-278. doi: 10.1016/j.jgg.2021.09.009. Epub 2021 Oct 28. J Genet Genomics. 2022. PMID: 34757037 Review.
-
Regulatory Functions of Protein Tyrosine Phosphatase Receptor Type O in Immune Cells.Front Immunol. 2021 Nov 22;12:783370. doi: 10.3389/fimmu.2021.783370. eCollection 2021. Front Immunol. 2021. PMID: 34880876 Free PMC article. Review.
Cited by
-
Liver Cell Mitophagy in Metabolic Dysfunction-Associated Steatotic Liver Disease and Liver Fibrosis.Antioxidants (Basel). 2024 Jun 15;13(6):729. doi: 10.3390/antiox13060729. Antioxidants (Basel). 2024. PMID: 38929168 Free PMC article. Review.
-
Therapeutic potential of targeting protein tyrosine phosphatases in liver diseases.Acta Pharm Sin B. 2024 Aug;14(8):3295-3311. doi: 10.1016/j.apsb.2024.05.006. Epub 2024 May 13. Acta Pharm Sin B. 2024. PMID: 39220870 Free PMC article. Review.
-
The Gut-Liver Axis in Chronic Liver Disease: A Macrophage Perspective.Cells. 2021 Oct 30;10(11):2959. doi: 10.3390/cells10112959. Cells. 2021. PMID: 34831182 Free PMC article. Review.
-
MiR-6869-5p Induces M2 Polarization by Regulating PTPRO in Gestational Diabetes Mellitus.Mediators Inflamm. 2021 Apr 30;2021:6696636. doi: 10.1155/2021/6696636. eCollection 2021. Mediators Inflamm. 2021. PMID: 34007244 Free PMC article.
-
Hepatic Macrophages as Targets for the MSC-Based Cell Therapy in Non-Alcoholic Steatohepatitis.Biomedicines. 2023 Nov 14;11(11):3056. doi: 10.3390/biomedicines11113056. Biomedicines. 2023. PMID: 38002056 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

