A New Hope for CD56negCD16pos NK Cells as Unconventional Cytotoxic Mediators: An Adaptation to Chronic Diseases
- PMID: 32373555
- PMCID: PMC7186373
- DOI: 10.3389/fcimb.2020.00162
A New Hope for CD56negCD16pos NK Cells as Unconventional Cytotoxic Mediators: An Adaptation to Chronic Diseases
Abstract
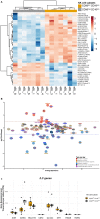
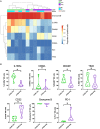
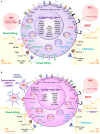
Natural Killer (NK) cells play an essential role in antiviral and anti-tumoral immune responses. In peripheral blood, NK cells are commonly classified into two major subsets: CD56brightCD16neg and CD56dimCD16pos despite the characterization of a CD56negCD16pos subset 25 years ago. Since then, several studies have described the prevalence of an CD56negCD16pos NK cell subset in viral non-controllers as the basis for their NK cell dysfunction. However, the mechanistic basis for their cytotoxic impairment is unclear. Recently, using a strict flow cytometry gating strategy to exclude monocytes, we reported an accumulation of CD56negCD16pos NK cells in Plasmodium falciparum malaria-exposed children and pediatric cancer patients diagnosed with endemic Burkitt lymphoma (eBL). Here, we use live-sorted cells, histological staining, bulk RNA-sequencing and flow cytometry to confirm that this CD56negCD16pos NK cell subset has the same morphological features as the other NK cell subsets and a similar transcriptional profile compared to CD56dimCD16pos NK cells with only 120 genes differentially expressed (fold change of 1.5, p < 0.01 and FDR<0.05) out of 9235 transcripts. CD56negCD16pos NK cells have a distinct profile with significantly higher expression of MPEG1 (perforin 2), FCGR3B (CD16b), FCGR2A, and FCGR2B (CD32A and B) as well as CD6, CD84, HLA-DR, LILRB1/2, and PDCD1 (PD-1), whereas Interleukin 18 (IL18) receptor genes (IL18RAP and IL18R1), cytotoxic genes such as KLRF1 (NKp80) and NCR1 (NKp46), and inhibitory HAVCR2 (TIM-3) are significantly down-regulated compared to CD56dimCD16pos NK cells. Together, these data confirm that CD56negCD16pos cells are legitimate NK cells, yet their transcriptional and protein expression profiles suggest their cytotoxic potential is mediated by pathways reliant on antibodies such as antibody-dependent cell cytotoxicity (ADCC), antibody-dependent respiratory burst (ADRB), and enhanced by complement receptor 3 (CR3) and FAS/FASL interaction. Our findings support the premise that chronic diseases induce NK cell modifications that circumvent proinflammatory mediators involved in direct cytotoxicity. Therefore, individuals with such altered NK cell profiles may respond differently to NK-mediated immunotherapies, infections or vaccines depending on which cytotoxic mechanisms are being engaged.
Keywords: CD56negCD16pos subset; endemic Burkitt lymphoma; epstein-barr virus; malaria; natural killer cells; transcription profile.
Copyright © 2020 Forconi, Oduor, Oluoch, Ong'echa, Münz, Bailey and Moormann.
Figures


References
-
- Andrews S. (2014). FastQC. A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

