Single-Cell Transcriptional Profiling of Cells Derived From Regenerating Alveolar Ducts
- PMID: 32373614
- PMCID: PMC7186418
- DOI: 10.3389/fmed.2020.00112
Single-Cell Transcriptional Profiling of Cells Derived From Regenerating Alveolar Ducts
Abstract
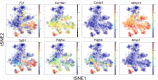
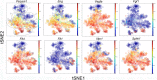
Lung regeneration occurs in a variety of adult mammals after surgical removal of one lung (pneumonectomy). Previous studies of murine post-pneumonectomy lung growth have identified regenerative "hotspots" in subpleural alveolar ducts; however, the cell-types participating in this process remain unclear. To identify the single cells participating in post-pneumonectomy lung growth, we used laser microdissection, enzymatic digestion and microfluidic isolation. Single-cell transcriptional analysis of the murine alveolar duct cells was performed using the C1 integrated fluidic circuit (Fluidigm) and a custom PCR panel designed for lung growth and repair genes. The multi-dimensional data set was analyzed using visualization software based on the tSNE algorithm. The analysis identified 6 cell clusters; 1 cell cluster was present only after pneumonectomy. This post-pneumonectomy cluster was significantly less transcriptionally active than 3 other clusters and may represent a transitional cell population. A provisional cluster identity for 4 of the 6 cell clusters was obtained by embedding bulk transcriptional data into the tSNE analysis. The transcriptional pattern of the 6 clusters was further analyzed for genes associated with lung repair, matrix production, and angiogenesis. The data demonstrated that multiple cell-types (clusters) transcribed genes linked to these basic functions. We conclude that the coordinated gene expression across multiple cell clusters is likely a response to a shared regenerative microenvironment within the subpleural alveolar ducts.
Keywords: aerobic glycolysis; cholangiocarcinoma; glucose metabolism; metabolic reprogramming; warburg effect.
Copyright © 2020 Ysasi, Bennett, Wagner, Valenzuela, Servais, Tsuda, Pyne, Li, Grimsby, Pokharel, Livak, Ackermann, Blainey and Mentzer.
Figures






Similar articles
-
Deformation-induced transitional myofibroblasts contribute to compensatory lung growth.Am J Physiol Lung Cell Mol Physiol. 2017 Jan 1;312(1):L79-L88. doi: 10.1152/ajplung.00383.2016. Epub 2016 Nov 11. Am J Physiol Lung Cell Mol Physiol. 2017. PMID: 27836901 Free PMC article.
-
Laser microdissection of the alveolar duct enables single-cell genomic analysis.Front Oncol. 2014 Sep 24;4:260. doi: 10.3389/fonc.2014.00260. eCollection 2014. Front Oncol. 2014. PMID: 25309876 Free PMC article.
-
Spatial dependence of alveolar angiogenesis in post-pneumonectomy lung growth.Angiogenesis. 2012 Mar;15(1):23-32. doi: 10.1007/s10456-011-9236-y. Epub 2011 Oct 4. Angiogenesis. 2012. PMID: 21969134 Free PMC article.
-
The alveolar surface network: a new anatomy and its physiological significance.Anat Rec. 1998 Aug;251(4):491-527. doi: 10.1002/(SICI)1097-0185(199808)251:4<491::AID-AR8>3.0.CO;2-V. Anat Rec. 1998. PMID: 9713987 Review.
-
Histochemical, Biochemical and Cell Biological aspects of tail regeneration in lizard, an amniote model for studies on tissue regeneration.Prog Histochem Cytochem. 2014 Jan;48(4):143-244. doi: 10.1016/j.proghi.2013.12.001. Epub 2014 Jan 1. Prog Histochem Cytochem. 2014. PMID: 24387878 Review.
Cited by
-
Alveologenesis: What Governs Secondary Septa Formation.Int J Mol Sci. 2021 Nov 9;22(22):12107. doi: 10.3390/ijms222212107. Int J Mol Sci. 2021. PMID: 34829987 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

