Genetic and expression variations of cell cycle pathway genes in brain tumor patients
- PMID: 32373934
- PMCID: PMC7225413
- DOI: 10.1042/BSR20190629
Genetic and expression variations of cell cycle pathway genes in brain tumor patients
Abstract
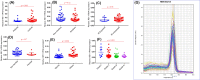
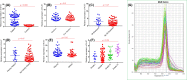
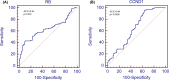
The present study was designed to determine the association between the genetic polymorphisms/expression variations of RB1 and CCND1 genes and brain tumor risk. For this purpose, 250 blood samples of brain tumor patients along with 250 controls (cohort I) and 96 brain tumor tissues (cohort II) with adjacent control section were collected. Mutation analysis of RB1 (rs137853294, rs121913300) and CCND1 (rs614367, rs498136) genes was performed using ARMS-PCR followed by sequencing, and expression analysis was performed using real-time PCR and immunohistochemistry. The results showed homozygous mutant genotype of RB1 gene polymorphism, rs121913300 (P=0.003) and CCND1 gene polymorphism rs614367 (P=0.01) were associated significantly with brain tumor risk. Moreover, significant down-regulation of RB1 (P=0.005) and up-regulation of CCND1 (P=0.0001) gene was observed in brain tumor sections vs controls. Spearman correlation showed significant negative correlation between RB1 vs proliferation marker, Ki-67 (r = -0.291*, P<0.05) in brain tumors. Expression levels of selected genes were also assessed at protein level using immunohistochemical analysis (IHC) and signification down-regulation of RB1 (P=0.0001) and up-regulation of CCND1 (P=0.0001) was observed in brain tumor compared with control sections. In conclusion, it is suggested that polymorphisms/expression variations of RB1 and CCND1 genes may be associated with increased risk of brain tumor.
Keywords: Brain tumor; CCND1; Expression analysis; Polymorphism; RB1.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

