Identification of Susceptibility Loci for Spontaneous Coronary Artery Dissection
- PMID: 32374345
- PMCID: PMC7203673
- DOI: 10.1001/jamacardio.2020.0872
Identification of Susceptibility Loci for Spontaneous Coronary Artery Dissection
Abstract
Importance: Spontaneous coronary artery dissection (SCAD), an idiopathic disorder that predominantly affects young to middle-aged women, has emerged as an important cause of acute coronary syndrome, myocardial infarction, and sudden cardiac death.
Objective: To identify common single-nucleotide variants (SNVs) associated with SCAD susceptibility.
Design, setting, and participants: This single-center genome-wide association study examined approximately 5 million genotyped and imputed SNVs and subsequent SNV-targeted replication analysis results in individuals enrolled in the Mayo Clinic SCAD registry from August 30, 2011, to August 2, 2018. Data analysis was performed from June 21, 2017, to December 30, 2019.
Main outcomes and measures: Genetic loci and positional candidate genes associated with SCAD.
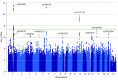
Results: This study included 484 white women with SCAD (mean [SD] age, 46.6 [9.2] years) and 1477 white female controls in the discovery cohort (mean [SD] age, 64.0 [14.5] years) and 183 white women with SCAD (mean [SD] age, 47.1 [9.9] years) and 340 white female controls in the replication cohort (mean [SD] age, 51.0 [15.3] years). Associations with SCAD risk reached genome-wide significance at 3 loci (1q21.3 [OR, 1.78; 95% CI, 1.51-2.09; P = 2.63 × 10-12], 6p24.1 [OR, 1.77; 95% CI, 1.51-2.09; P = 7.09 × 10-12], and 12q13.3 [OR, 1.67; 95% CI, 1.42-1.97; P = 3.62 × 10-10]), and 7 loci had evidence suggestive of an association (1q24.2 [OR, 2.10; 95% CI, 1.58-2.79; P = 2.88 × 10-7], 3q22.3 [OR, 1.47; 95% CI, 1.26-1.71; P = 6.65 × 10-7], 4q34.3 [OR, 1.84; 95% CI, 1.44-2.35; P = 9.80 × 10-7], 8q24.3 [OR, 2.57; 95% CI, 1.76-3.75; P = 9.65 × 10-7], 15q21.1 [OR, 1.75; 95% CI, 1.40-2.18; P = 7.23 × 10-7], 16q24.1 [OR, 1.91; 95% CI, 1.49-2.44; P = 2.56 × 10-7], and 21q22.11 [OR, 2.11; 95% CI, 1.59-2.82; P = 3.12 × 10-7]) after adjusting for the top 5 principal components. Associations were validated for 5 of the 10 risk alleles in the replication cohort. In a meta-analysis of the discovery and replication cohorts, associations for the 5 SNVs were significant, with relatively large effect sizes (1q21.3 [OR, 1.77; 95% CI, 1.54-2.03; P = 3.26 × 10-16], 6p24.1 [OR, 1.71; 95% CI, 1.49-1.97; P = 4.59 × 10-14], 12q13.3 [OR, 1.69; 95% CI, 1.47-1.94; P = 1.42 × 10-13], 15q21.1 [OR, 1.79; 95% CI, 1.48-2.17; P = 2.12 × 10-9], and 21q22.11 [OR, 2.18; 95% CI, 1.70-2.81; P = 1.09 × 10-9]). Each index SNV was within or near a gene highly expressed in arterial tissue and previously linked to SCAD (PHACTR1) and/or other vascular disorders (LRP1, LINC00310, and FBN1).
Conclusions and relevance: This study revealed 5 replicated risk loci and positional candidate genes for SCAD, most of which are associated with extracoronary arteriopathies. Moreover, the alternate alleles of 3 SNVs have been previously associated with atherosclerotic coronary artery disease, further implicating allelic susceptibility to coronary artery atherosclerosis vs dissection.
Conflict of interest statement
Figures

References
-
- Hayes SN, Kim ESH, Saw J, et al. ; American Heart Association Council on Peripheral Vascular Disease; Council on Clinical Cardiology; Council on Cardiovascular and Stroke Nursing; Council on Genomic and Precision Medicine; and Stroke Council . Spontaneous coronary artery dissection: current state of the science: a scientific statement from the American Heart Association. Circulation. 2018;137(19):e523-e557. doi:10.1161/CIR.0000000000000564 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

