Orotidine 5'-Monophosphate Decarboxylase: The Operation of Active Site Chains Within and Across Protein Subunits
- PMID: 32374983
- PMCID: PMC7476526
- DOI: 10.1021/acs.biochem.0c00241
Orotidine 5'-Monophosphate Decarboxylase: The Operation of Active Site Chains Within and Across Protein Subunits
Abstract
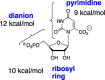
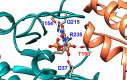
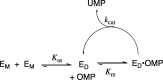
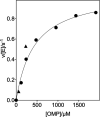
The D37 and T100' side chains of orotidine 5'-monophosphate decarboxylase (OMPDC) interact with the C-3' and C-2' ribosyl hydroxyl groups, respectively, of the bound substrate. We compare the intra-subunit interactions of D37 with the inter-subunit interactions of T100' by determining the effects of the D37G, D37A, T100'G, and T100'A substitutions on the following: (a) kcat and kcat/Km values for the OMPDC-catalyzed decarboxylations of OMP and 5-fluoroorotidine 5'-monophosphate (FOMP) and (b) the stability of dimeric OMPDC relative to the monomer. The D37G and T100'A substitutions resulted in 2 kcal mol-1 increases in ΔG† for kcat/Km for the decarboxylation of OMP, while the D37A and T100'G substitutions resulted in larger 4 and 5 kcal mol-1 increases, respectively, in ΔG†. The D37G and T100'A substitutions both resulted in smaller 2 kcal mol-1 decreases in ΔG† for the decarboxylation of FOMP compared to that of OMP. These results show that the D37G and T100'A substitutions affect the barrier to the chemical decarboxylation step while the D37A and T100'G substitutions also affect the barrier to a slow, ligand-driven enzyme conformational change. Substrate binding induces the movement of an α-helix (G'98-S'106) toward the substrate C-2' ribosyl hydroxy bound at the main subunit. The T100'G substitution destabilizes the enzyme dimer by 3.5 kcal mol-1 compared to the monomer, which is consistent with the known destabilization of α-helices by the internal Gly side chains [Serrano, L., et al. (1992) Nature, 356, 453-455]. We propose that the T100'G substitution weakens the α-helical contacts at the dimer interface, which results in a decrease in the dimer stability and an increase in the barrier to the ligand-driven conformational change.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

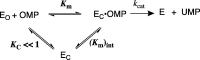


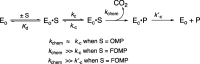


References
-
- Miller B. G.; Hassell A. M.; Wolfenden R.; Milburn M. V.; Short S. A. (2000) Anatomy of a proficient enzyme: the structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc. Natl. Acad. Sci. U. S. A. 97, 2011–2016. 10.1073/pnas.030409797. - DOI - PMC - PubMed
-
- Tsang W.-Y.; Wood B. M.; Wong F. M.; Wu W.; Gerlt J. A.; Amyes T. L.; Richard J. P. (2012) Proton Transfer from C-6 of Uridine 5′-Monophosphate Catalyzed by Orotidine 5′-Monophosphate Decarboxylase: Formation and Stability of a Vinyl Carbanion Intermediate and the Effect of a 5-Fluoro Substituent. J. Am. Chem. Soc. 134, 14580–14594. 10.1021/ja3058474. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

