The molecular basis of selective DNA binding by the BRG1 AT-hook and bromodomain
- PMID: 32376391
- PMCID: PMC7350285
- DOI: 10.1016/j.bbagrm.2020.194566
The molecular basis of selective DNA binding by the BRG1 AT-hook and bromodomain
Abstract
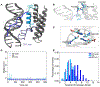
The ATP-dependent BAF chromatin remodeling complex plays a critical role in gene regulation by modulating chromatin architecture, and is frequently mutated in cancer. Indeed, subunits of the BAF complex are found to be mutated in >20% of human tumors. The mechanism by which BAF properly navigates chromatin is not fully understood, but is thought to involve a multivalent network of histone and DNA contacts. We previously identified a composite domain in the BRG1 ATPase subunit that is capable of associating with both histones and DNA in a multivalent manner. Mapping the DNA binding pocket revealed that it contains several cancer mutations. Here, we utilize SELEX-seq to investigate the DNA specificity of this composite domain and NMR spectroscopy and molecular modelling to determine the structural basis of DNA binding. Finally, we demonstrate that cancer mutations in this domain alter the mode of DNA association.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


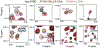


References
-
- Gatchalian J, Malik S, Ho J, Lee D-S, Kelso TWR, Shokhirev MN, Dixon JR, Hargreaves DC, A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells., Nature Communications. 9 (2018) 5139. doi:10.1038/s41467-018-07528-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

