Transcriptomics analysis of pericytes from retinas of diabetic animals reveals novel genes and molecular pathways relevant to blood-retinal barrier alterations in diabetic retinopathy
- PMID: 32376470
- PMCID: PMC7323486
- DOI: 10.1016/j.exer.2020.108043
Transcriptomics analysis of pericytes from retinas of diabetic animals reveals novel genes and molecular pathways relevant to blood-retinal barrier alterations in diabetic retinopathy
Abstract
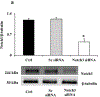
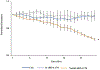
Selective pericyte loss, the histological hallmark of early diabetic retinopathy (DR), enhances the breakdown of the blood-retinal barrier (BRB) in diabetes. However, the role of pericytes on BRB alteration in diabetes and the signaling pathways involved in their effects are currently unknown. To understand the role of diabetes-induced molecular alteration of pericytes, we performed transcriptomic analysis of sorted retinal pericytes from mice model of diabetes. Retinal tissue from non-diabetic and diabetic (duration 3 months) mouse eyes (n = 10 in each group) were used to isolate pericytes through fluorescent activated cell sorting (FACS) using pericyte specific fluorescent antibodies, PDGFRb-APC. For RNA sequencing and qPCR analysis, a cDNA library was generated using template switching oligo and the resulting libraries were sequenced using paired-end Illumina sequencing. Molecular functional pathways were analyzed using differentially expressed genes (DEGs). Differential expression analysis revealed 217 genes significantly upregulated and 495 genes downregulated, in pericytes isolated from diabetic animals. These analyses revealed a core set of differentially expressed genes that could potentially contribute to the pericyte dysfunction in diabetes and highlighted the pattern of functional connectivity between key candidate genes and blood retinal barrier alteration mechanisms. The top up-regulated gene list included: Ext2, B3gat3, Gpc6, Pip5k1c and Pten and down-regulated genes included: Notch3, Xbp1, Gpc4, Atp1a2 and AKT3. Out of these genes, we further validated one of the down regulated genes, Notch 3 and its role in BRB alteration in diabetic retinopathy. We confirmed the downregulation of Notch3 expression in human retinal pericytes exposed to Advanced Glycation End-products (AGEs) treatment mimicking the chronic hyperglycemia effect. Exploration of pericyte-conditioned media demonstrated that loss of NOTCH3 in pericyte led to increased permeability of endothelial cell monolayers. Collectively, we identify a role for NOTCH3 in pericyte dysfunction in diabetes. Further validation of other DEGs to identify cell specific molecular change through whole transcriptomic approach in diabetic retina will provide novel insight into the pathogenesis of DR and novel therapeutic targets.
Keywords: Blood retinal barrier; Diabetic retinopathy; Notch3; Pericytes; RNA Sequencing.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing financial interests.
Figures




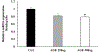

References
-
- Armulik A, Abramsson A, Betsholtz C, 2005. Endothelial/pericyte interactions. Circ Res 97, 512–523. - PubMed
-
- Chasseigneaux S, Moraca Y, Cochois-Guegan V, Boulay AC, Gilbert A, Le Crom S, Blugeon C, Firmo C, Cisternino S, Laplanche JL, Curis E, Decleves X, Saubamea B, 2018. Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci Rep 8, 12272. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

