Medium-throughput Drug Screening of Patient-derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy
- PMID: 32376656
- PMCID: PMC8366292
- DOI: 10.1158/1078-0432.CCR-20-0073
Medium-throughput Drug Screening of Patient-derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy
Abstract
Purpose: Patients with colorectal cancer with peritoneal metastases (CRPMs) have limited treatment options and the lowest colorectal cancer survival rates. We aimed to determine whether organoid testing could help guide precision treatment for patients with CRPMs, as the clinical utility of prospective, functional drug screening including nonstandard agents is unknown.
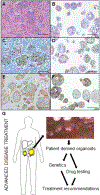
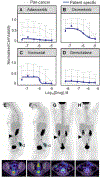
Experimental design: CRPM organoids (peritonoids) isolated from patients underwent parallel next-generation sequencing and medium-throughput drug panel testing ex vivo to identify specific drug sensitivities for each patient. We measured the utility of such a service including: success of peritonoid generation, time to cultivate peritonoids, reproducibility of the medium-throughput drug testing, and documented changes to clinical therapy as a result of the testing.
Results: Peritonoids were successfully generated and validated from 68% (19/28) of patients undergoing standard care. Genomic and drug profiling was completed within 8 weeks and a formal report ranking drug sensitivities was provided to the medical oncology team upon failure of standard care treatment. This resulted in a treatment change for two patients, one of whom had a partial response despite previously progressing on multiple rounds of standard care chemotherapy. The barrier to implementing this technology in Australia is the need for drug access and funding for off-label indications.
Conclusions: Our approach is feasible, reproducible, and can guide novel therapeutic choices in this poor prognosis cohort, where new treatment options are urgently needed. This platform is relevant to many solid organ malignancies.
©2020 American Association for Cancer Research.
Conflict of interest statement
Figures


References
-
- Franko J, Shi Q, Goldman C, Pockaj B, Nelson G, Goldberg R, et al. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: a pooled analysis of north central cancer treatment group phase III trials N9741 and N9841. J Clin Oncol 2012;30(3):263–7 doi 10.1200/JCO.2011.37.1039. - DOI - PMC - PubMed
-
- Franko J, Shi Q, Meyers JP, Maughan TS, Adams RA, Seymour MT, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol 2016;17(12):1709–19 doi 10.1016/S1470-2045(16)30500-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

