Polyamines regulate gene expression by stimulating translation of histone acetyltransferase mRNAs
- PMID: 32376690
- PMCID: PMC7324502
- DOI: 10.1074/jbc.RA120.013833
Polyamines regulate gene expression by stimulating translation of histone acetyltransferase mRNAs
Abstract
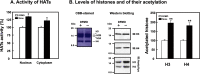
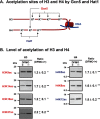
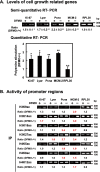
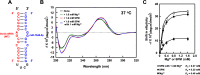
Polyamines regulate gene expression in Escherichia coli by translationally stimulating mRNAs encoding global transcription factors. In this study, we focused on histone acetylation, one of the mechanisms of epigenetic regulation of gene expression, to attempt to clarify the role of polyamines in the regulation of gene expression in eukaryotes. We found that activities of histone acetyltransferases in both the nucleus and cytoplasm decreased significantly in polyamine-reduced mouse mammary carcinoma FM3A cells. Although protein levels of histones H3 and H4 did not change in control and polyamine-reduced cells, acetylation of histones H3 and H4 was greatly decreased in the polyamine-reduced cells. Next, we used control and polyamine-reduced cells to identify histone acetyltransferases whose synthesis is stimulated by polyamines. We found that polyamines stimulate the translation of histone acetyltransferases GCN5 and HAT1. Accordingly, GCN5- and HAT1-catalyzed acetylation of specific lysine residues on histones H3 and H4 was stimulated by polyamines. Consistent with these findings, transcription of genes required for cell proliferation was enhanced by polyamines. These results indicate that polyamines regulate gene expression by enhancing the expression of the histone acetyltransferases GCN5 and HAT1 at the level of translation. Mechanistically, polyamines enhanced the interaction of microRNA-7648-5p (miR-7648-5p) with the 5'-UTR of GCN5 mRNA, resulting in stimulation of translation due to the destabilization of the double-stranded RNA (dsRNA) between the 5'-UTR and the ORF of GCN5 mRNA. Because HAT1 mRNA has a short 5'-UTR, polyamines may enhance initiation complex formation directly on this mRNA.
Keywords: gene expression; histone acetylase; histone acetyltransferase; histone acetyltransferase 1 (HAT1); histone acetyltransferase GCN5 (GCN5); histone modification; miR-7648-5p; polyamine; polyamine modulon; spermidine; translation regulation; translational regulation.
© 2020 Sakamoto et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

