Circulating Tumor DNA Using Tagged Targeted Deep Sequencing to Assess Minimal Residual Disease in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy
- PMID: 32377196
- PMCID: PMC7196957
- DOI: 10.1155/2020/8132507
Circulating Tumor DNA Using Tagged Targeted Deep Sequencing to Assess Minimal Residual Disease in Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy
Abstract
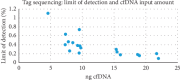
In breast cancer patients undergoing neoadjuvant chemotherapy before surgery, there is an unmet need for noninvasive predictive biomarkers of response. The analysis of circulating tumor DNA (ctDNA) in particular has been the object of several reports, but few of them have studied the applicability of tagged targeted deep sequencing (tTDS) to clinical practice and its performance compared with droplet digital PCR (ddPCR). Here, we present the first results from an ongoing study involving a prospectively accrued, monocentric cohort of patients affected by invasive breast cancer, undergoing neoadjuvant chemotherapy followed by surgery with curative intent as per clinical practice. A pretreatment tumor biopsy and plasma samples were collected before and during treatment, after surgery, and every six months henceforth or until relapse, whichever came first. Pretreatment biopsies were sequenced with a 409-gene massive parallel sequencing (MPS) panel, allowing the identification of target mutations and their research in plasma by tTDS and ddPCR as a complementary approach. Using tTDS, we demonstrated the presence of at least one deleterious mutation in all the relapsed cases we studied (n = 4), with an average lead time of six months before clinical relapse. The association with ddPCR was suboptimal, and only one relapsed patient could be identified with such method. tTDS shows potential as an early noninvasive method for the detection of MRD in BC patients.
Copyright © 2020 Gabriella Cirmena et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures



References
LinkOut - more resources
Full Text Sources

