The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant
- PMID: 32377354
- PMCID: PMC7192901
- DOI: 10.1038/s41438-020-0288-2
The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant
Abstract
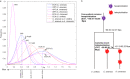
Tea is one of the most popular nonalcoholic beverages due to its characteristic secondary metabolites with numerous health benefits. Although two draft genomes of tea plant (Camellia sinensis) have been published recently, the lack of chromosome-scale assembly hampers the understanding of the fundamental genomic architecture of tea plant and potential improvement. Here, we performed a genome-wide chromosome conformation capture technique (Hi-C) to obtain a chromosome-scale assembly based on the draft genome of C. sinensis var. sinensis and successfully ordered 2984.7 Mb (94.7%) scaffolds into 15 chromosomes. The scaffold N50 of the improved genome was 218.1 Mb, ~157-fold higher than that of the draft genome. Collinearity comparison of genome sequences and two genetic maps validated the high contiguity and accuracy of the chromosome-scale assembly. We clarified that only one Camellia recent tetraploidization event (CRT, 58.9-61.7 million years ago (Mya)) occurred after the core-eudicot common hexaploidization event (146.6-152.7 Mya). Meanwhile, 9243 genes (28.6%) occurred in tandem duplication, and most of these expanded after the CRT event. These gene duplicates increased functionally divergent genes that play important roles in tea-specific biosynthesis or stress response. Sixty-four catechin- and caffeine-related quantitative trait loci (QTLs) were anchored to chromosome assembly. Of these, two catechin-related QTL hotspots were derived from the CRT event, which illustrated that polyploidy has played a dramatic role in the diversification of tea germplasms. The availability of a chromosome-scale genome of tea plant holds great promise for the understanding of genome evolution and the discovery of novel genes contributing to agronomically beneficial traits in future breeding programs.
Keywords: Comparative genomics; Genome evolution.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures





References
-
- Chen, L., Apostolides, Z. & Chen, Z. M. Global Tea Breeding: Achievements, Challenges and Perspectives (University Press-Springer, Hangzhou, Zhejiang; 2012).
-
- Chen L, Yao MZ, Wang XC, Yang YJ. Tea genetic resources in China. Int. J. Tea Sci. 2012;8(2):55–64.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

