Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing
- PMID: 32377375
- PMCID: PMC7197635
- DOI: 10.1038/s41421-020-0168-9
Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing
Erratum in
-
Erratum: Author Correction: Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing.Cell Discov. 2020 Jun 20;6:41. doi: 10.1038/s41421-020-00187-5. eCollection 2020. Cell Discov. 2020. PMID: 32595980 Free PMC article.
Abstract
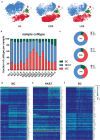
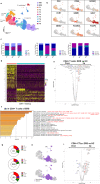
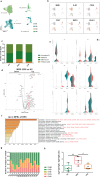
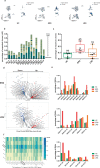
COVID-19, caused by SARS-CoV-2, has recently affected over 1,200,000 people and killed more than 60,000. The key immune cell subsets change and their states during the course of COVID-19 remain unclear. We sought to comprehensively characterize the transcriptional changes in peripheral blood mononuclear cells during the recovery stage of COVID-19 by single-cell RNA sequencing technique. It was found that T cells decreased remarkably, whereas monocytes increased in patients in the early recovery stage (ERS) of COVID-19. There was an increased ratio of classical CD14++ monocytes with high inflammatory gene expression as well as a greater abundance of CD14++IL1β+ monocytes in the ERS. CD4+ T cells and CD8+ T cells decreased significantly and expressed high levels of inflammatory genes in the ERS. Among the B cells, the plasma cells increased remarkably, whereas the naïve B cells decreased. Several novel B cell-receptor (BCR) changes were identified, such as IGHV3-23 and IGHV3-7, and isotypes (IGHV3-15, IGHV3-30, and IGKV3-11) previously used for virus vaccine development were confirmed. The strongest pairing frequencies, IGHV3-23-IGHJ4, indicated a monoclonal state associated with SARS-CoV-2 specificity, which had not been reported yet. Furthermore, integrated analysis predicted that IL-1β and M-CSF may be novel candidate target genes for inflammatory storm and that TNFSF13, IL-18, IL-2, and IL-4 may be beneficial for the recovery of COVID-19 patients. Our study provides the first evidence of an inflammatory immune signature in the ERS, suggesting COVID-19 patients are still vulnerable after hospital discharge. Identification of novel BCR signaling may lead to the development of vaccines and antibodies for the treatment of COVID-19.
Keywords: Immunology; Mechanisms of disease.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures


References
-
- Livingston, E., Bucher, K. Coronavirus disease 2019 (COVID-19) in Italy. JAMA10.1001/jama.2020.4344 (2020). - PubMed
-
- World Health Organization. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). WHO. https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-... (2020).
Grants and funding
- 81830054, 81988101,91859205/National Natural Science Foundation of China (National Science Foundation of China)
- 81722034/National Natural Science Foundation of China (National Science Foundation of China)
- 81670015/National Natural Science Foundation of China (National Science Foundation of China)
- 81530028,81721003/National Natural Science Foundation of China (National Science Foundation of China)
- 2018ZX09101002, 2017ZX100203205/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

