Long noncoding RNA ANRIL promotes the malignant progression of cholangiocarcinoma by epigenetically repressing ERRFI1 expression
- PMID: 32378752
- PMCID: PMC7385372
- DOI: 10.1111/cas.14447
Long noncoding RNA ANRIL promotes the malignant progression of cholangiocarcinoma by epigenetically repressing ERRFI1 expression
Abstract
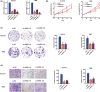
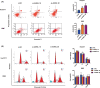
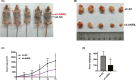
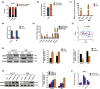
Long noncoding RNAs (lncRNAs) have recently been verified to have significant regulatory functions in many types of human cancers. The lncRNA ANRIL is transcribed from the INK4b-ARF-INK4a gene cluster in the opposite direction. Whether ANRIL can act as an oncogenic molecule in cholangiocarcinoma (CCA) remains unknown. Our data show that ANRIL knockdown greatly inhibited CCA cell proliferation and migration in vitro and in vivo. According to the results of RNA sequencing analysis, ANRIL knockdown dramatically altered target genes associated with the cell cycle, cell proliferation, and apoptosis. By binding to a component of the epigenetic modification complex enhancer of zeste homolog 2 (EZH2), ANRIL could maintain lysine residue 27 of histone 3 (H3K27me3) levels in the promoter of ERBB receptor feedback inhibitor 1 (ERRFI1), which is a tumor suppressor gene in CCA. In this way, ERRFI1 expression was suppressed in CCA cells. These data verified the key role of the epigenetic regulation of ANRIL in CCA oncogenesis and indicate its potential as a target for CCA intervention.
Keywords: ANRIL; ERRFI1; cholangiocarcinoma; epigenetic regulation; long noncoding RNA.
© 2020 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
None.
Figures


References
-
- Zhu AX. Future directions in the treatment of cholangiocarcinoma. Best Pract Res Clin Gastroenterol. 2015;29:355‐361. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

