Quantifying antibiotic impact on within-patient dynamics of extended-spectrum beta-lactamase resistance
- PMID: 32379042
- PMCID: PMC7205461
- DOI: 10.7554/eLife.49206
Quantifying antibiotic impact on within-patient dynamics of extended-spectrum beta-lactamase resistance
Abstract
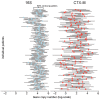
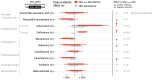
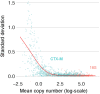
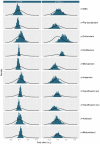
Antibiotic-induced perturbation of the human gut flora is expected to play an important role in mediating the relationship between antibiotic use and the population prevalence of antibiotic resistance in bacteria, but little is known about how antibiotics affect within-host resistance dynamics. Here we develop a data-driven model of the within-host dynamics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae. We use blaCTX-M (the most widespread ESBL gene family) and 16S rRNA (a proxy for bacterial load) abundance data from 833 rectal swabs from 133 ESBL-positive patients followed up in a prospective cohort study in three European hospitals. We find that cefuroxime and ceftriaxone are associated with increased blaCTX-M abundance during treatment (21% and 10% daily increase, respectively), while treatment with meropenem, piperacillin-tazobactam, and oral ciprofloxacin is associated with decreased blaCTX-M (8% daily decrease for all). The model predicts that typical antibiotic exposures can have substantial long-term effects on blaCTX-M carriage duration.
Trial registration: ClinicalTrials.gov NCT01208519.
Keywords: antibiotic resistance; epidemiology; extended-spectrum beta-lactamase; global health; gut microbiota; human; infectious disease; microbiology; resistance carriage; state-space model; within-host dynamics.
Plain language summary
Bacteria that are resistant to antibiotics are a growing global health crisis. One type of antibiotic resistance arises when certain bacteria that can produce enzymes called extended-spectrum beta-lactamases (or ESBLs for short) become more common in the gut. These enzymes stop important antibiotics, like penicillin, from working. However, exactly which antibiotics and treatment durations contribute to the emergence of this antibiotic resistance remain unknown. Now, Niehus et al. find certain antibiotics that are associated with an increase in the number of gut bacteria carrying antibiotic resistance genes for ESBL enzymes. First, rectal swabs collected from 133 patients from three European hospitals were analysed to measure the total gut bacteria and the number of genes for ESBL enzymes. These samples had been collected at several time points including when the patient was first admitted to hospital, then every two to three days during their stay, and finally when they were discharged. Combining the analysis of the samples with details of the patients’ charts showed that treatment with two antibiotics: cefuroxime and ceftriaxone, was linked to an increase in ESBL genes in the gut bacteria. Other antibiotics – namely, meropenem, piperacillin-tazobactam and oral ciprofloxacin – were associated with a decrease in the number of bacteria with ESBL genes. Niehus et al. then performed further analysis to see if different treatment regimens affected how long patients were carrying gut bacteria with ESBL genes. This predicted that a longer course of meropenem, 14 days rather than 5 days, would shorten the length of time patients carried ESBL-resistant bacteria in their guts by 70%, although this effect will likely depend on the location of the hospital and the local prevalence of other types of antibiotic resistance. This analysis reveals new details about how antibiotic treatment can affect ESBL resistance genes. More studies are needed to understand how antibiotics affect other antibiotic resistance genes and how resistant bacteria spread. This will help scientists understand how much specific antibiotic regimens contribute to antibiotic resistance. It may also help scientists develop new antibiotic treatment strategies that reduce antibiotic resistance.
© 2020, Niehus et al.
Conflict of interest statement
RN, Ev, YM, AT, CL, YC, HG, ET, BC, LP, SM No competing interests declared, BC Reviewing Editor, eLife
Figures






References
-
- Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, Grobusch MP, Lashof AMO, Molhoek N, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. The Lancet Infectious Diseases. 2017;17:78–85. doi: 10.1016/S1473-3099(16)30319-X. - DOI - PubMed
-
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. Definitions and guidelines for research on antibiotic persistence. Nature Reviews Microbiology. 2019;17:441–448. doi: 10.1038/s41579-019-0196-3. - DOI - PMC - PubMed
-
- Buelow E, Gonzalez TB, Versluis D, Oostdijk EA, Ogilvie LA, van Mourik MS, Oosterink E, van Passel MW, Smidt H, D'Andrea MM, de Been M, Jones BV, Willems RJ, Bonten MJ, van Schaik W. Effects of selective digestive decontamination (SDD) on the gut resistome. Journal of Antimicrobial Chemotherapy. 2014;69:2215–2223. doi: 10.1093/jac/dku092. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

