Biomolecular complex viewed by dynamic nuclear polarization solid-state NMR spectroscopy
- PMID: 32379300
- PMCID: PMC7565284
- DOI: 10.1042/BST20191084
Biomolecular complex viewed by dynamic nuclear polarization solid-state NMR spectroscopy
Abstract
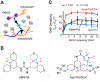
Solid-state nuclear magnetic resonance (ssNMR) is an indispensable tool for elucidating the structure and dynamics of insoluble and non-crystalline biomolecules. The recent advances in the sensitivity-enhancing technique magic-angle spinning dynamic nuclear polarization (MAS-DNP) have substantially expanded the territory of ssNMR investigations and enabled the detection of polymer interfaces in a cellular environment. This article highlights the emerging MAS-DNP approaches and their applications to the analysis of biomolecular composites and intact cells to determine the folding pathway and ligand binding of proteins, the structural polymorphism of low-populated biopolymers, as well as the physical interactions between carbohydrates, proteins, and lignin. These structural features provide an atomic-level understanding of many cellular processes, promoting the development of better biomaterials and inhibitors. It is anticipated that the capabilities of MAS-DNP in biomolecular and biomaterial research will be further enlarged by the rapid development of instrumentation and methodology.
Keywords: cell wall; dynamic nuclear polarization; membrane proteins; pathogenic fungi; polysaccharides; solid-state NMR.
© 2020 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Figures


References
-
- McDermott A (2009) Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Annu. Rev. Biophys 38, 385–403 - PubMed
-
- Meier BH, Riek R and Bockmann A (2017) Emerging Structural Understanding of Amyloid Fibrils by Solid-State NMR. Trends Biochem. Sci 42, 777–787 - PubMed
-
- Comellas G and Rienstra CM (2013) Protein Structure Determination by Magic-Angle Spinning Solid-State NMR, and Insights into the Formation, Structure, and Stability of Amyloid Fibrils. Annu. Rev. Biophys 42, 515–536 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

