Molecular detection of SARS-CoV-2 in formalin-fixed, paraffin-embedded specimens
- PMID: 32379723
- PMCID: PMC7406253
- DOI: 10.1172/jci.insight.139042
Molecular detection of SARS-CoV-2 in formalin-fixed, paraffin-embedded specimens
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of human coronavirus disease 2019 (COVID-19), emerged in Wuhan, China, in December 2019. The virus rapidly spread globally, resulting in a public health crisis including almost 5 million cases and 323,256 deaths as of May 21, 2020. Here, we describe the identification and evaluation of commercially available reagents and assays for the molecular detection of SARS-CoV-2 in infected FFPE cell pellets. We identified a suitable rabbit polyclonal anti-SARS-CoV spike protein antibody and a mouse monoclonal anti-SARS-CoV nucleocapsid protein (NP) antibody for cross-detection of the respective SARS-CoV-2 proteins by IHC and immunofluorescence assay (IFA). Next, we established RNAscope in situ hybridization (ISH) to detect SARS-CoV-2 RNA. Furthermore, we established a multiplex FISH (mFISH) to detect positive-sense SARS-CoV-2 RNA and negative-sense SARS-CoV-2 RNA (a replicative intermediate indicating viral replication). Finally, we developed a dual staining assay using IHC and ISH to detect SARS-CoV-2 antigen and RNA in the same FFPE section. It is hoped that these reagents and assays will accelerate COVID-19 pathogenesis studies in humans and in COVID-19 animal models.
Keywords: COVID-19; Molecular diagnosis; Molecular pathology; Virology.
Conflict of interest statement
Figures


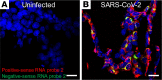
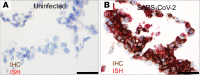
Update of
-
Molecular Detection of SARS-CoV-2 in Formalin Fixed Paraffin Embedded Specimens.bioRxiv [Preprint]. 2020 Apr 21:2020.04.21.042911. doi: 10.1101/2020.04.21.042911. bioRxiv. 2020. Update in: JCI Insight. 2020 Jun 18;5(12):139042. doi: 10.1172/jci.insight.139042. PMID: 32511350 Free PMC article. Updated. Preprint.
References
-
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ Accessed May 21, 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

