Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures
- PMID: 32379750
- PMCID: PMC7237046
- DOI: 10.1371/journal.pcbi.1007852
Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures
Abstract
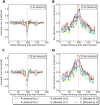
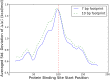
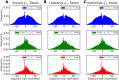
Single nucleotide polymorphisms are widely associated with disease, but the ways in which they cause altered phenotypes are often unclear, especially when they appear in non-coding regions. One way in which non-coding polymorphisms could cause disease is by affecting crucial RNA-protein interactions. While it is clear that changing a protein binding motif will alter protein binding, it has been shown that single nucleotide polymorphisms can affect RNA secondary structure, and here we show that single nucleotide polymorphisms can affect RNA-protein interactions from outside binding motifs through altered RNA secondary structure. By using a modified version of the Vienna Package and PAR-CLIP data for HuR (ELAVL1) in humans we characterize the genome-wide effect of single nucleotide polymorphisms on HuR binding and show that they can have a many-fold effect on the affinity of HuR binding to RNA transcripts from tens of bases away. We also find some evidence that the effect of single nucleotide polymorphisms on protein binding might be under selection, with the non-reference alleles tending to make it harder for a protein to bind.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

