Transcriptome analysis identifies genes involved with the development of umbilical hernias in pigs
- PMID: 32379844
- PMCID: PMC7205231
- DOI: 10.1371/journal.pone.0232542
Transcriptome analysis identifies genes involved with the development of umbilical hernias in pigs
Abstract
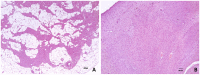
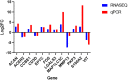
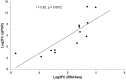
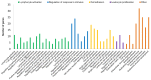
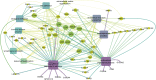
Umbilical hernia (UH) is one of the most frequent defects affecting pig production, however, it also affects humans and other mammals. UH is characterized as an abnormal protrusion of the abdominal contents to the umbilical region, causing pain, discomfort and reduced performance in pigs. Some genomic regions associated to UH have already been identified, however, no study involving RNA sequencing was performed when umbilical tissue is considered. Therefore, here, we have sequenced the umbilical ring transcriptome of five normal and five UH-affected pigs to uncover genes and pathways involved with UH development. A total of 13,216 transcripts were expressed in the umbilical ring tissue. From those, 230 genes were differentially expressed (DE) between normal and UH-affected pigs (FDR <0.05), being 145 downregulated and 85 upregulated in the affected compared to the normal pigs. A total of 68 significant biological processes were identified and the most relevant were extracellular matrix, immune system, anatomical development, cell adhesion, membrane components, receptor activation, calcium binding and immune synapse. The results pointed out ACAN, MMPs, COLs, EPYC, VIT, CCBE1 and LGALS3 as strong candidates to trigger umbilical hernias in pigs since they act in the extracellular matrix remodeling and in the production, integrity and resistance of the collagen. We have generated the first transcriptome of the pig umbilical ring tissue, which allowed the identification of genes that had not yet been related to umbilical hernias in pigs. Nevertheless, further studies are needed to identify the causal mutations, SNPs and CNVs in these genes to improve our understanding of the mechanisms of gene regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Alexandratos N, Bruinsma J. World Agriculture towards 2030/2050: the 2012 revision. WORLD Agric. 2012.
-
- Searcy-Bernal R, Gardner IA, Hird DW. Effects of and factors associated with umbilical hernias in a swine herd. J Am Vet Med Assoc. 1994. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

