The HMGB1-RAGE axis modulates the growth of autophagy-deficient hepatic tumors
- PMID: 32382012
- PMCID: PMC7206028
- DOI: 10.1038/s41419-020-2536-7
The HMGB1-RAGE axis modulates the growth of autophagy-deficient hepatic tumors
Abstract
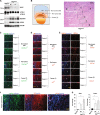
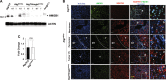
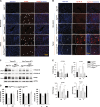
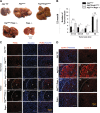
Autophagy is an intracellular lysosomal degradative pathway important for tumor surveillance. Autophagy deficiency can lead to tumorigenesis. Autophagy is also known to be important for the aggressive growth of tumors, yet the mechanism that sustains the growth of autophagy-deficient tumors is not unclear. We previously reported that progression of hepatic tumors developed in autophagy-deficient livers required high mobility group box 1 (HMGB1), which was released from autophagy-deficient hepatocytes. In this study we examined the pathological features of the hepatic tumors and the mechanism of HMGB1-mediated tumorigenesis. We found that in liver-specific autophagy-deficient (Atg7ΔHep) mice the tumors cells were still deficient in autophagy and could also release HMGB1. Histological analysis using cell-specific markers suggested that fibroblast and ductular cells were present only outside the tumor whereas macrophages were present both inside and outside the tumor. Genetic deletion of Hmgb1 or one of its receptors, receptor for advanced glycated end product (Rage), retarded liver tumor development. HMGB1 and RAGE enhanced the proliferation capability of the autophagy-deficient hepatocytes and tumors. However, RAGE expression was only found on ductual cells and Kupffer's cells but not on hepatoctyes, suggesting that HMGB1 might promote hepatic tumor growth through a paracrine mode, which altered the tumor microenvironment. Finally, RNAseq analysis of the tumors indicated that HMGB1 induced a much broad changes in tumors. In particular, genes related to mitochondrial structures or functions were enriched among those differentially expressed in tumors in the presence or absence of HMGB1, revealing a potentially important role of mitochondria in sustaining the growth of autophagy-deficient liver tumors via HMGB1 stimulation.
Conflict of interest statement
The authors declared that they have no conflict of interest.
Figures


Similar articles
-
HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers.J Clin Invest. 2018 Jun 1;128(6):2419-2435. doi: 10.1172/JCI91814. Epub 2018 May 7. J Clin Invest. 2018. PMID: 29558368 Free PMC article.
-
High Mobility Group Box-1 Drives Fibrosis Progression Signaling via the Receptor for Advanced Glycation End Products in Mice.Hepatology. 2018 Dec;68(6):2380-2404. doi: 10.1002/hep.30093. Epub 2018 Nov 13. Hepatology. 2018. PMID: 29774570 Free PMC article.
-
Involvement of the nuclear high mobility group B1 peptides released from injured hepatocytes in murine hepatic fibrogenesis.Biochim Biophys Acta. 2014 Sep;1842(9):1720-32. doi: 10.1016/j.bbadis.2014.06.017. Epub 2014 Jun 23. Biochim Biophys Acta. 2014. PMID: 24970745
-
Role of High-Mobility Group Box-1 in Liver Pathogenesis.Int J Mol Sci. 2019 Oct 25;20(21):5314. doi: 10.3390/ijms20215314. Int J Mol Sci. 2019. PMID: 31731454 Free PMC article. Review.
-
The Immune Tolerance Role of the HMGB1-RAGE Axis.Cells. 2021 Mar 5;10(3):564. doi: 10.3390/cells10030564. Cells. 2021. PMID: 33807604 Free PMC article. Review.
Cited by
-
Neutrophil Extracellular Traps (NETs) and Damage-Associated Molecular Patterns (DAMPs): Two Potential Targets for COVID-19 Treatment.Mediators Inflamm. 2020 Jul 16;2020:7527953. doi: 10.1155/2020/7527953. eCollection 2020. Mediators Inflamm. 2020. PMID: 32724296 Free PMC article. Review.
-
Programmed Cell Death Tunes Tumor Immunity.Front Immunol. 2022 Mar 30;13:847345. doi: 10.3389/fimmu.2022.847345. eCollection 2022. Front Immunol. 2022. PMID: 35432318 Free PMC article. Review.
-
Crosstalk between cGAS-STING pathway and autophagy in cancer immunity.Front Immunol. 2023 Mar 1;14:1139595. doi: 10.3389/fimmu.2023.1139595. eCollection 2023. Front Immunol. 2023. PMID: 36936940 Free PMC article. Review.
-
Emerging Roles of High-mobility Group Box-1 in Liver Disease.J Clin Transl Hepatol. 2024 Dec 28;12(12):1043-1056. doi: 10.14218/JCTH.2024.00317. Epub 2024 Oct 22. J Clin Transl Hepatol. 2024. PMID: 39649031 Free PMC article. Review.
-
Discovery of Novel Host Molecular Factors Underlying HBV/HCV Infection.Front Cell Dev Biol. 2021 Aug 12;9:690882. doi: 10.3389/fcell.2021.690882. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34458256 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

